研究チーム
Team研究チーム
研究項目A02 計画研究:革新的ユビキチンケモテクノロジーの創出
ユビキチンコードのケミカル合成
研究代表者 岡本 晃充東京大学 大学院工学系研究科化学生命工学専攻 教授 |
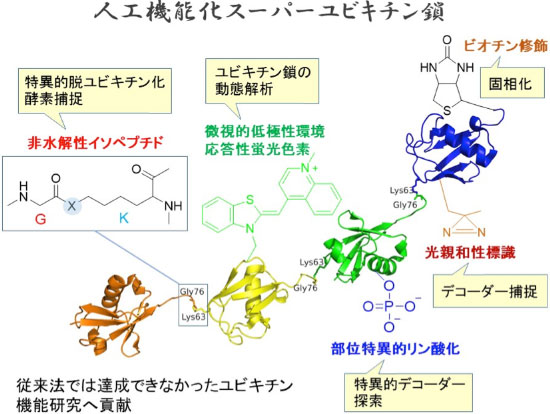
本計画研究の概要
ユビキチンコードとスーパーユビキチンを創り、それらを利用することでユビキチンコードの作動機構を解明します。まず、ユビキチンの化学合成法を確立し、様々なユビキチン鎖と人工機能化ユビキチンの合成に展開します。続いて、合成ユビキチン鎖を用いて、デコーダー分子の機能解析やユビキチン鎖の構造・動態解析を実施します。
1. ユビキチンの化学合成法の確立
ユビキチンを2〜3のフラグメントに分割して、それらのフラグメントペプチドの合成、ケミカルライゲーションによるフラグメントの連結、ライゲーションに用いられたシステインの脱硫反応によってユビキチンを化学合成します。
2. 多様なユビキチン鎖のコンビナトリアル化学合成
特殊ε-Lys-Glyを適切に導入したフラグメントをあらかじめ用意しておくことによって、多様なポリユビキチン鎖をコンビナトリアルに化学合成します。次いで、リン酸化ユビキチン鎖やアセチル化ユビキチン鎖、複雑なユビキチンコードであるK48/K63分岐鎖やK63/M1混合鎖の合成に着手します。化学合成したユビキチン鎖をベイトとして特異的なデコーダー分子を捕捉して、最新型質量分析計を用いて同定します。
3. 人工機能化ユビキチン(スーパーユビキチン)の合成
蛍光色素や光親和性標識などを部位特異的に導入したユビキチンを作製します。さらには、ビオチン修飾末端を有するユビキチンや脱ユビキチン化酵素耐性ユビキチンの合成などを検討します。作出したスーパーユビキチンを細胞に導入し、細胞内局在や細胞応答を解析します。
4. スーパーユビキチン鎖の物性解析ならびにユビキチンコードの形成解析
合成ポリユビキチン鎖の構造解析および結合力解析を進めるとともに、特異環境で蛍光発光するユビキチンを用いて、ユビキチンコード形成を蛍光でモニタリングする系を開発します。
5. ユビキチン1分子観察
ポリユビキチン鎖内・鎖間で2種類の異なる蛍光標識を導入して、蛍光相互相関分光法を用いることにより、試験管中ならびに細胞中での1分子レベルのユビキチンの動態および他分子との結合の様子をモニタリングします。
研究概要を示す模式図
本領域での研究成果
- Sueoka T, Koyama K, Hayashi G, *Okamoto A.
Chemistry-driven Epigenetic Investigation of Histone and DNA Modifications.
Chem. Rec. 18, 1727-1744 (2018)
PMID: 30070422 - Kamo N, *Hayashi G, *Okamoto A.
Triple Function of 4-Mercaptophenylacetic Acid Promotes One-Pot Multiple Peptide Ligation.
Angew. Chem. Int. Ed. 57, 16533-16537 (2018)
PMID: 30346110 - *Hayashi G, Yanase M, Nakatsuka Y, *Okamoto A.
Simultaneous and Traceless Ligation of Peptide Fragments on DNA Scaffold.
Biomacromolecules 20, 1246-1253 (2019)
PMID: 30677290 - Yanase M, Nakatsu K, Cardos C J, Konda Y, *Hayashi G, *Okamoto A.
Cysteinylprolyl Imide (CPI) Peptide: A Highly Reactive and Easily Accessible Crypto-thioester for Chemical Protein Synthesis.
Chem. Sci. 10, 5967-5975 (2019)
PMID: 31360403 - Kamo N, *Hayashi G, *Okamoto A.
Chemical Synthesis of Cys-Containing Protein via Chemoselective Deprotection with Different Palladium Complexes.
Org. Lett. 21, 8378-8382 (2019)
PMID: 31560553 - Izuta S, *Yamaguchi S, Kosaka T, *Okamoto A.
Reversible and Photoresponsive Immobilization of Nonadherent Cells by Spiropyran-Conjugated PEG-Lipids.
ACS Appl. Bio Mater. 2, 33-38 (2019) - *Yamaguchi S, Higashi K, Azuma T, *Okamoto A.
Supramolecular Polymeric Hydrogels for Ultrasound-guided Protein Release.
Biotechnol. J. 14, 1800530 (2019)
PMID: 30810275 - Usami K, Xiao K, *Okamoto A.
Efficient Ketose Production by Hydroxyapatite Catalyst in a Continuous Flow Module.
ACS Sustainable Chem. Eng. 7, 3372-3377 (2019) - Jarzebska N A, *Yamaguchi S, Izuta S, Kosaka T, Yamahira S, Nagamune T, *Okamoto A.
Photo-responsive materials with strong cell trapping ability for light-guided manipulation of nonadherent cells.
Biomater. Sci. 7, 4514-4518 (2019)
PMID: 31602442 - Morihiro K, *Okamoto A.
A highly constrained nucleic acid analog based on α-L-threosamine.
Nucleosides Nucleotides Nucleic Acids 39, 270-279 (2020)
PMID: 31530088 - Chen J, Morihiro K, Fukui D, Guo L, *Okamoto A.
Live‐Cell Sensing of Telomerase Activity Using Hybridization-Sensitive Fluorescent Oligonucleotide Probes.
ChemBioChem 21, 1022-1027 (2020)
PMID: 31840916 - Nakatsu K, Yanase M, *Hayashi G, *Okamoto A.
Fmoc-Compatible and C-terminal-Sequence-Independent Peptide Alkyl Thioester Formation Using Cysteinylprolyl Imide.
Org. Lett. 22, 4670–4674 (2020)
PMID: 32484687 - *Yamaguchi S, Takagi R, Hosogane T, Ohashi Y, Sakai Y, Sakakihara S, Iino R, Tabata K, Noji H, *Okamoto A.
Single cell array enclosed with a photodegradable hydrogel in microwells for image-based cell classification and selective photorelease of cells.
ACS Appl. Bio Mater. 3, 5887–5895 (2020) - Ogata M, Hayashi G, Ichiu A, *Okamoto A.
L-DNA-tagged fluorescence in situ hybridization for highly sensitive imaging of RNAs in single cells.
Org. Biomol. Chem. 18, 8084-8088 (2020)
PMID: 33001106 - Hao F, Murphy KJ, Kujirai T, Kamo N, Kato J, Koyama M, Okamoto A, Hayashi G, Kurumizaka H, *Hayes JJ.
Acetylation-Modulated Communication between the H3 N-Terminal Tail Domain and the Intrinsically Disordered H1 C-Terminal Domain.
Nucleic Acids Res. 48, 11510–11520 (2020)
PMID: 33125082 - Morihiro K, Ishinabe T, Takatsu M, Osumi H, Osawa T, *Okamoto A.
Floxuridine Oligomers Activated under Hypoxic Environment.
J. Am. Chem. Soc. 143, 3340–3347 (2021)
PMID: 33648338 - Kamo N, Kujirai T, Kurumizaka H, Murakami H, *Hayashi G, *Okamoto A.
Organoruthenium-Catalyzed Chemical Protein Synthesis to Elucidate the Functions of Epigenetic Modifications on Heterochromatin Factors.
Chem. Sci. 12, 5926-5937 (2021) - Kamo N, *Hayashi G, *Okamoto A.
Silyl-Protected Propargyl Glycine for Multiple Labeling of Peptides by Chemoselective Silyl-Deprotection.
Tetrahedron Lett. 73, 153093 (2021) - Koyama K, Hayashi G, Ueda H, Ota S, Nagae G, Aburatani H, *Okamoto A.
Base-Resolution Analysis of 5-Hydroxymethylcytidine by Selective Oxidation and Reverse Transcription Arrest.
Org. Biomol. Chem. 19, 6478-6486 (2021)
PMID: 34241617 - Morihiro K, Moriyama Y, Nemoto Y, Osumi H, *Okamoto A.
anti-syn Unnatural Base Pair Enables Alphabet-Expanded DNA Self-Assembly.
J. Am. Chem. Soc. 143, 14207-14217 (2021) - Pan M, Zorbas C, Sugaya M, Ishiguro K, Kato M, Nishida M, Zhang H-F, Candeias M M, Okamoto A, Ishikawa T, Soga T, Aburatani H, Sakai J, Matsumura Y, Suzuki T, Proud C G, Lafontaine D L J, *Osawa T.
Glutamine Deficiency in Solid Tumor Cells Confers Resistance to Ribosomal RNA Synthesis Inhibitors.
Nat. Commun. 13, 3706 (2022)
PMID: 35764642 - Yamahira S, Misawa R, Kosaka T, Tan M, Izuta S, Yamashita H, Heike Y, Okamoto A, Nagamune T, *Yamaguchi S.
Photoactivatable Materials for Versatile Single-Cell Patterning Based on the Photocaging of Cell-Anchoring Moieties through Lipid Self-Assembly.
J. Am. Chem. Soc. 144, 13154-13162 (2022)
PMID: 35767880 - Nakatsu K, Okamoto A, Hayashi G, *Murakami H.
Repetitive Thiazolidine Deprotection Using a Thioester-Compatible Aldehyde Scavenger for One-Pot Multiple Peptide Ligation.
Angew. Chem. Int. Ed. 61, e202206240 (2022)
PMID: 35881031 - *Yamaguchi S, Ikeda R, Umeda Y, Kosaka T, Yamahira S, *Okamoto A.
Chemoenzymatic Labeling to Visualize Intercellular Contacts Using Lipidated Sortase A.
ChemBioChem 23, e202200474 (2022)
PMID: 35976800 - Kosaka T, *Yamaguchi S, Izuta S, Yamahira S, Shibasaki Y, Tateno H, *Okamoto A.
Bioorthogonal Photoreactive Surfaces for Single-Cell Analysis of Intercellular Communications.
J. Am. Chem. Soc. 144, 17980-17988 (2022)
PMID: 36126284 - *Yamaguchi S, Yamamoto K, Yamamoto R, Takamori S, Ishiwatari A, Minamihata K, Nagamune T, *Okamoto A.
Intracellular Protein Photoactivation Using Sterically Bulky Caging.
ChemBioChem 23, e202200476 (2022)
PMID: 36173993 - Akizuki Y, Morita M, Mori Y, Kaiho-Soma A, Dixit S, Endo A, Shimogawa M, Hayashi G, Naito M, Okamoto A, Tanaka K, Saeki Y, *Ohtake F.
cIAP1-based degraders induce degradation via branched ubiquitin architectures.
Nat Chem Biol. 19, 311-322 (2023)
PMID: 36316570 - *Morihiro K, Osumi H, Morita S, Hattori T, Baba M, Harada N, Ohashi R, *Okamoto A.
Oncolytic Hairpin DNA Pair: Selective Cytotoxic Inducer through MicroRNA-Triggered DNA Self-Assembly.
J. Am. Chem. Soc. 145, 135-142 (2023)
PMID: 36538570 - Liu S, Morihiro K, Takeuchi F, Li Y, *Okamoto A.
Interstrand Crosslinking Oligonucleotides Elucidate the Effect of Metal Ions on the Methylation Status of Repetitive DNA Elements.
Front. Chem. 11, 1122474 (2023)
PMID: 36711237
関連する代表的な論文
- *Okamoto A.
DNA-Osmium Complexes: Recent Developments in the Operative Chemical Analysis of DNA Epigenetic Modification.
ChemMedChem 9, 1958-1965 (2014)
PMID: 24989699 - Hayashi G, Sakamoto R, *Okamoto A.
2-Oxazoline Formation for Selective Chemical Labeling of 5-Hydroxylysine.
Chem. Asian J. 10, 1138-1141 (2015)
PMID: 25757225 - Yamamoto K, Chikaoka Y, Hayashi G, Sakamoto R, Yamamoto R, Sugiyama A, Kodama T, Okamoto A, *Kawamura T.
Middle-Down and Chemical Proteomic Approaches to Reveal Histone H4 Modification Dynamics in Cell Cycle: Label-Free Semi-Quantification of Histone Tail Peptide Modifications Including Phosphorylation and Highly Sensitive Capture of Histone PTM Binding Proteins Using Photo-Reactive Crosslinkers.
Mass Spectrom. 4, A0039 (2015)
PMID: 26819910 - Hayashi G, Sueoka T, *Okamoto A.
In vitro and in cell analysis of chemically synthesized histone H2A with multiple modifications.
Chem. Commun. 52, 4999-5002 (2016)
PMID: 26981611 - Hayashi G, Koyama K, Shiota H, Kamio A, Umeda T, Nagae G, Aburatani H, *Okamoto A.
Base-Resolution Analysis of 5-Hydroxymethylcytosine by One-Pot Bisulfite-Free Chemical Conversion with Peroxotungstate.
J. Am. Chem. Soc. 138, 14178-14181 (2016)
PMID: 27766835 - Yamaguchi S, Matsushita T, Izuta S, Katada S, Ura M, Ikeda T, Hayashi G, Suzuki Y, Kobayashi Y, Tokunaga K, Ozeki Y, *Okamoto A.
Chemically-activatable alkyne-tagged probe for imaging microdomains in lipid bilayer membranes.
Sci. Rep. 7, 41007 (2017)
PMID: 28117375 - Matsushita T, Moriyama Y, Nagae G, Aburatani H, *Okamoto A.
DNA-friendly Cu(II)/TEMPO-catalyzed 5-hydroxymethylcytosine-specific oxidation.
Chem. Commun. 53, 5756-5759 (2017)
PMID: 28492666 - Hayashi G, Kamo N, *Okamoto A.
Chemical synthesis of dual labeled protein via differently protected alkynes enables intramolecular FRET analysis.
Chem. Commun. 53, 5918-5921 (2017)
PMID: 28513662 - Guo L, *Okamoto A.
Fluorescence-switching RNA for detection of bacterial ribosome.
Chem. Commun. 53, 9406-9409 (2017)
PMID: 28765840 - Sueoka T, Hayashi G, *Okamoto A.
Regulation of the Stability of the Histone H2A-H2B Dimer by H2A Tyr57 Phosphorylation.
Biochemistry 56, 4767-4772 (2017)
PMID: 28813589





