Atsuya Nishiyama
Mechanistic studies of ubiquitin signaling at DNA methylation sites using chemo-technology
 |
Atsuya Nishiyama, PhDDivision of Cancer Cell Biology, The Institute of Medical Science, The University of Tokyo |
|---|
Research summary
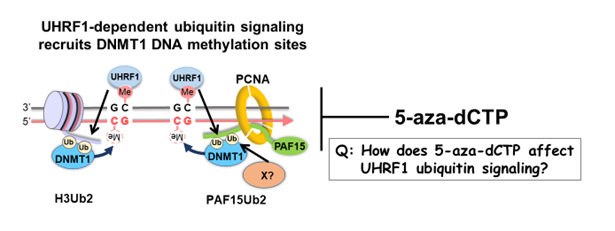
DNA methylation is one of the best-known epigenetic modifications and regulates various biological events, including transposon repression and transcriptional regulation through changes in chromatin structure. Hemi-methylated DNA binding protein UHRF1 E3 ubiquitin ligase, and the DNA methyltransferase DNMT1 plays a pivotal role in maintaining DNA methylation patterns during cell proliferation. Recent work by ourselves and other groups has shown that multiple mono-ubiquitylation by UHRF1 targeting histone H3 and PAF15 is critical for DNA maintenance methylation. However, the full set of factors involved in these processes remains unknown. In this research project, we aim to elucidate the molecular mechanism of ubiquitin-dependent maintenance DNA methylation by combining chemotechnologies and in vitro chromosome replication systems capable of reproducing the process of DNA methylation maintenance. In particular, we aim to clarify how 5-aza-dCTP, a DNMT1 inhibitor, and an anticancer agent, affects maintenance DNA machinery. To this end, we will employ a technique called chromatin mass spectrometry (CHROMASS) to monitor protein assembly and disassembly on 5-aza-dCTP-containing chromatin systematically. We will also work on the search for novel DNMT1 and UHRF1 inhibitors.
Publications
- Mishima Y, Brueckner L, Takahashi S, Kawakami T, Otani J, Shinohara A, Takeshita K, Garvilles RG, Watanabe M, Sakai N, Takeshima H, Nachtegael C, Nishiyama A, Nakanishi M, Arita K, Nakashima K, Hojo H, Suetake I.
Enhanced processivity of Dnmt1 by monoubiquitinated histone H3.
Genes to Cells 25, 22-32 (2020)
PMID: 31680384 - *Nishiyama A, Mulholland C, Bultmann S, Kori A, Endo A, Saeki Y, Qin W, Trummer C, Chiba Y, Yokoyama H, Kumamoto S, Kawakami T, Hojo H, Nagae G, Aburatani H, Tanaka K, *Arita K, *Leonhardt H, *Nakanishi M.
Two distinct modes of DNMT1 recruitment ensure the stable maintenance DNA methylation.
Nat. Commun. 11, 1222 (2020)
PMID: 32144273
Faculty Opinions - Mulholland CB, Nishiyama A, Ryan J, Nakamura R, Yiğit M, Glück IM, Trummer C, Qin W, Bartoschek MD, Traube FR, Parsa E, Ugur E, Modic M, Acharya A, Stolz P, Ziegenhain C, Wierer M, Enard W, Carell T, Lamb DC, Takeda H, Nakanishi M, Bultmann S, *Leonhardt H.
Recent evolution of a TET-controlled and DPPA3/STELLA-driven pathway of passive DNA demethylation in mammals.
Nat. Commun. 11, 5972 (2020)
PMID: 33235224
Former Publications
- *Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, Koseki H, *Nakanishi M.
Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication.
Nature 502,249-253 (2013)
PMID: 24013172 - Nishiyama A, Yamaguchi L, *Nakanishi M.
Regulation of maintenance DNA methylation via histone ubiquitylation.
J. Biochem. 159,9-15 (2016)
PMID: 26590302 - Misaki T, Yamaguchi L, Sun J, Orii M, *Nishiyama A, *Nakanishi M.
The replication foci targeting sequence (RFTS) of DNMT1 functions as a potent histone H3 binding domain regulated by autoinhibition.
Biochem. Biophys. Res. Commun. 470,741-747 (2016)
PMID: 26774338 - Yamaguchi L, *Nishiyama A, Misaki T, Johmura Y, Ueda J, Arita K, Nagao K, Obuse C, *Nakanishi M.
Usp7-dependent histone H3 deubiquitylation regulates maintenance of DNA methylation.
Sci. Rep. 7,55 (2017)
PMID: 28246399 - Ishiyama S, Nishiyama A, Saeki Y, Moritsugu K, Morimoto D, Yamaguchi L, Arai N, Matsumura R, Kawakami T, Mishima Y, Hojo H, Shimamura S, Ishikawa F, Tajima S,Tanaka K, Ariyoshi M, Shirakawa M, Ikeguchi M, Kidera A, *Suetake I, *Arita K, *Nakanishi M.
Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance.
Mol. Cell 68,350-360(2017)
PMID: 29053958