Takuya Terai
Identification of ubiquitin ligase substrates and development of protein degradation method using evolutionary molecular engineering
 |
Takuya Terai, Ph.DBiomolecular Chemistry Laboratory, Department of Chemistry, School of Science, The University of Tokyo |
|---|
Research summary
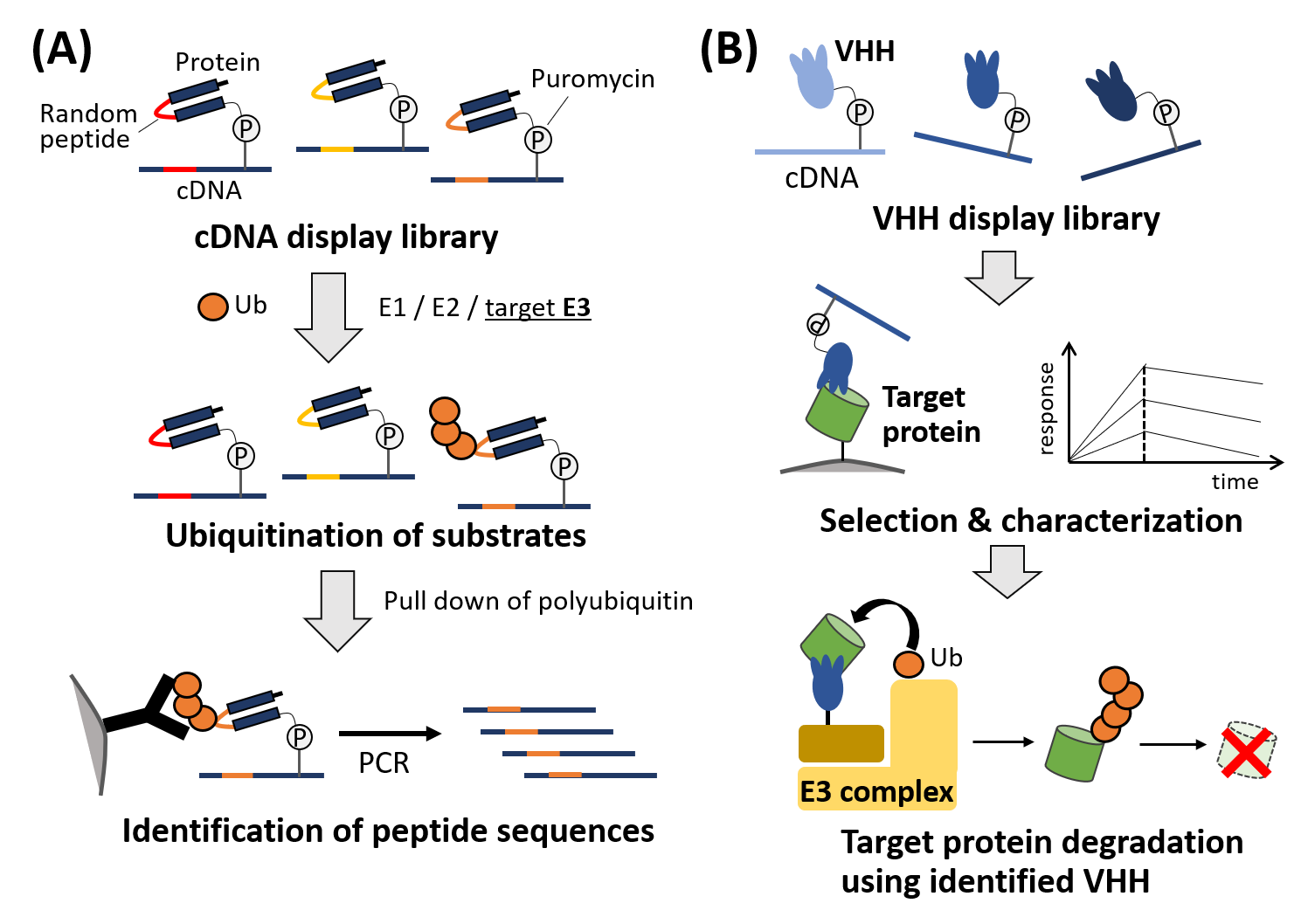
Although protein ubiquitination plays a variety of roles in cells, the knowledge on substrate specificity of many ubiquitin ligases (E3) in our body is still very limited. To address this issue, I will perform large-scale screening of peptide sequences that are selectively recognized and degraded by the E3 ligase of interest, using the power of in vitro display technology (cDNA display) of peptides. cDNA display is a single molecule conjugate of a peptide and its corresponding cDNA, which has mainly been used for in vitro selection of peptide aptamers for target proteins. In this study, a cDNA display library incorporating random peptide moiety is prepared and incubated with E3 and other components for in vitro ubiquitination. Ubiquitinated cDNA display molecules are selectively collected using anti-ubiquitin antibody, and the attached DNA sequences are amplified and analyzed by deep sequencing (Figure 1, left). With this technique, we will be able to investigate global substrate preferences of the target E3 enzyme. Such information will be useful to identify endogenous protein targets as well as to develop a method of targeted protein degradation under specific conditions.
In this project, I also plan to identify novel VHHs (single domain antibodies derived from heavy chain antibodies in camelid) that bind to specific proteins and apply them to targeted protein degradation. Protein degradation technologies such as PROTACs and SNIPERs are attracting huge attention in drug discovery, but it is not always easy to find a specific small molecule binder for the target protein. VHHs are smaller than standard IgG antibodies and compatible with intracellular environment. Also, protein engineering and in vitro selection of VHHs are relatively easy. Hence, as the second project of this research, we will screen VHHs for the target proteins using cDNA display technology. Then, using the identified VHHs as a reagent that recruit the target to E3, a novel protein knock-down strategy will be developed (Figure 1, right). Finally, we plan to combine this method with reported light/small molecule sensitive VHH technologies, to achieve conditional degradation of target proteins in cells.
Publications
- Anzai H, *Terai T, Wakabayashi-Nakao K, Noguchi T, Kumachi S, Tsuchiya M, *Nemoto N.
Interleukin-17A Peptide Aptamers with an Unexpected Binding Moiety Selected by cDNA Display under Heterogenous Conditions.
ACS Med. Chem. Lett. 12, 1427-1434 (2021)
PMID: 34531951 - Zhu W, Takeuchi S, Imai S, Terada T, Ueda T, Nasu Y, *Terai T, *Campbell R E.
Chemigenetic indicators based on synthetic chelators and green fluorescent protein.
Nat. Chem. Biol. 19, 38-44 (2023)
PMID: 36138142
Former Publications
- Kobayashi S, Terai T, Yoshikawa Y, Ohkawa R, Ebihara M, Hayashi M, Takiguchi K, *Nemoto N.
In vitro selection of random peptides against artificial lipid bilayers: a potential tool to immobilize molecules on membranes.
Chem. Commun. 53, 3458–3461 (2017)
PMID: 28271115 - *Terai T, Anzai H, *Nemoto N.
Selection of Peptides that Associate with Dye-Conjugated Solid Surfaces in a pH-Dependent Manner Using cDNA Display.
ACS Omega 4, 7378-7384 (2019)
PMID: 31459836 - Jayathilake C, Kumachi S, Arai H, Motohashi M, Terai T, Murakami A, *Nemoto N.
In vitro selection of anti-gliadin single-domain antibodies from a naïve library for cDNA-display mediated immuno-PCR.
Anal. Biochem. 589, 113490 (2020)
PMID: 31678363 - *Terai T, Koike T, *Nemoto N.
Photocrosslinking of cDNA Display Molecules with Their Target Proteins as a New Strategy for Peptide Selection.
Molecules 25, 1472 (2020)
PMID: 32214008