Atsuya Nishiyama
Deciphering ubiquitin code using chemo-technologies and advanced proteomics
 |
Atsuya Nishiyama, PhDDivision of Cancer Cell Biology, The Institute of Medical Science, The University of Tokyo |
|---|
Research summary
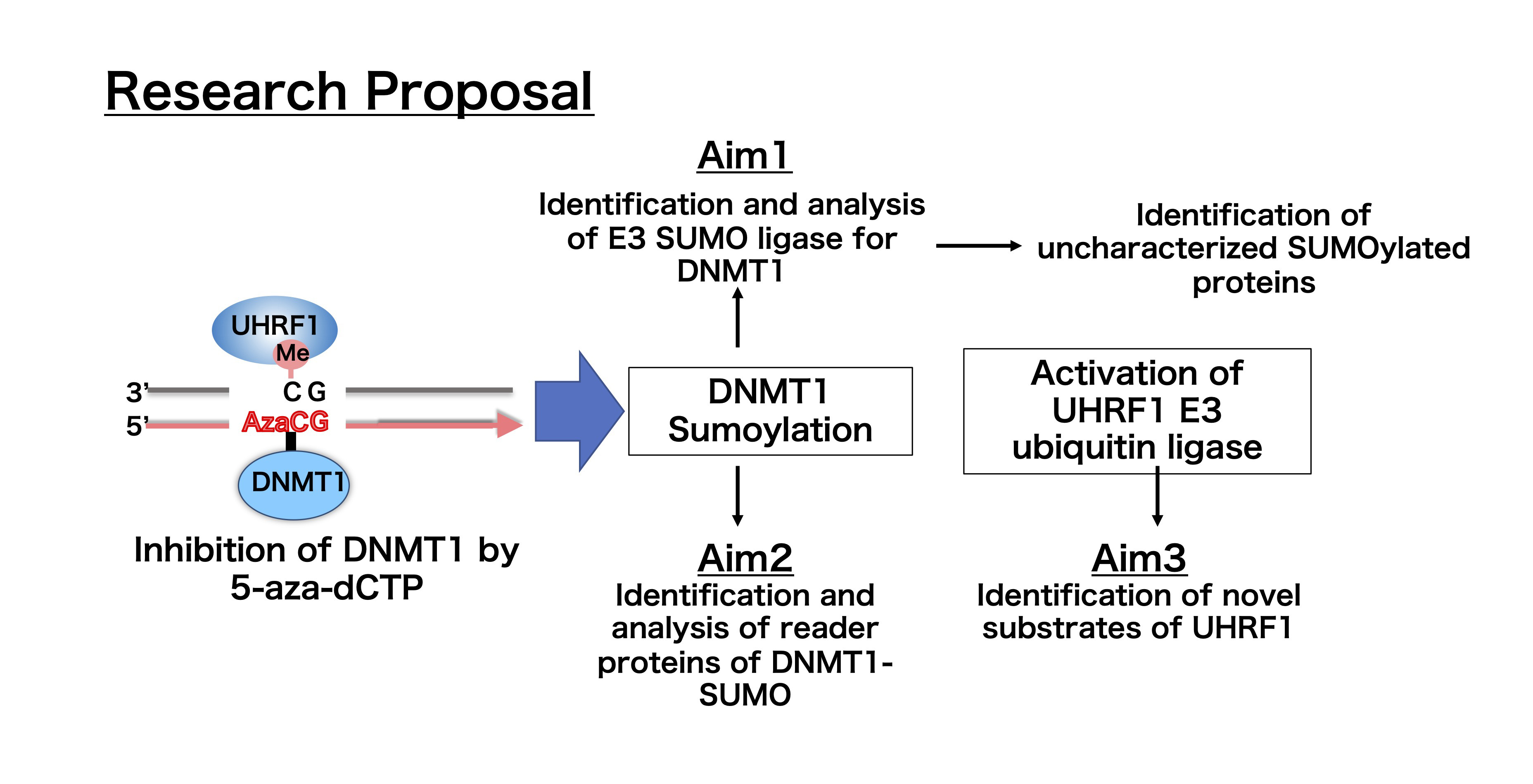
DNA methylation is an epigenetic chemical modification that plays an essential role in a variety of biological processes. In recent years, DNA methyltransferase inhibitors that induce DNA hypomethylation have become a promising tool in cancer therapy. 5-aza-2-deoxycytidine (5-aza-2-dC), a derivative of deoxycytidine, is incorporated into genomic DNA during DNA replication and traps DNMT1 in the genome by forming irreversible covalent bonds. DNMT1 is then removed from the cell by proteasome-dependent proteolysis. However, the molecular mechanism of DNA-DNMT1 cross-link repair remains unclear. Using a cell-free system derived from Xenopus egg extracts, we found that 5-aza-dCTP induces a high level of DNMT1 Sumoylation and activation of UHRF1 E3 ubiquitin ligase. Based on these data, we propose the central hypothesis that DNMT1 Sumoylation plays a critical role in regulating DNMT1-DNA cross-link repair. Our specific aims are outlined below:
- Identification and analysis of E3 SUMO ligase for DNMT1
- Identification and analysis of reader proteins of Sumoylated DNMT1
- Identification of novel substrates of UHRF1
Publications
- Kumamoto S, *Nishiyama A, Chiba Y, Miyashita R, Konishi C, Azuma Y, *Nakanishi M.
HPF1-dependent PARP activation promotes LIG3-XRCC1-mediated backup pathway of Okazaki fragment ligation.
Nucleic Acids Research 49, 5003-5016 (2021)
PMID: 33872376 - Kori S, Shibahashi Y, Ekimoto T, Nishiyama A, Yoshimi S, Yamaguchi K, Nagatoishi S, Ohta M, Tsumoto K, Nakanishi M, Defossez PA, Ikeguchi M, *Arita K.
Structure-based screening combined with computational and biochemical analyses identified the inhibitor targeting the binding of DNA Ligase 1 to UHRF1.
Bioorg Med. Chem. 52, 116500 (2021)
PMID: 34801826 - Kikuchi A, Onoda H, Yamaguchi K, Kori S, Matsuzawa S, Chiba Y, Tanimoto S, Yoshimi S, Sato H, Yamagata A, Shirouzu M, Adachi N, Sharif J, Koseki H, Nishiyama A, Nakanishi M, Defossez PA , Arita K.
Structural basis for activation of DNMT1.
Nature Communications 13, 7130 (2022)
PMID: 36414620 - Hata K, Kobayashi N, Sugimura K, Qin W, Haxholli D, Chiba Y, Yoshimi S, Hayashi G, Onoda H, Ikegami T, Mulholland CB, Nishiyama A, Nakanishi M, Leonhardt H, Konuma T, Arita K.
Structural basis for the unique multifaceted interaction of DPPA3 with the UHRF1 PHD finger.
Nucleic Acids Res. 50, 12527-12542 (2022)
PMID: 36420895 - Miyashita R, *Nishiyama A, Qin W, Chiba Y, Kori S, Kato N, Konishi C, Kumamoto S, Kozuka-Hata H, Oyama M, Kawasoe Y, Tsurimoto T, Takahashi TS, Leonhardt H, Arita K, *Nakanishi M.
The termination of UHRF1-dependent PAF15 ubiquitin signaling is regulated by USP7 and ATAD5.
eLife. AOP (2023)
PMID: 36734974
Former Publications
- *Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, Koseki H, *Nakanishi M.
Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication.
Nature 502,249-253 (2013)
PMID: 24013172 - Nishiyama A, Yamaguchi L, *Nakanishi M.
Regulation of maintenance DNA methylation via histone ubiquitylation.
J. Biochem. 159,9-15 (2016)
PMID: 26590302 - Yamaguchi L, *Nishiyama A, Misaki T, Johmura Y, Ueda J, Arita K, Nagao K, Obuse C, *Nakanishi M.
Usp7-dependent histone H3 deubiquitylation regulates maintenance of DNA methylation.
Sci. Rep. 7,55 (2017)
PMID: 28246399 - Ishiyama S, Nishiyama A, Saeki Y, Moritsugu K, Morimoto D, Yamaguchi L, Arai N, Matsumura R, Kawakami T, Mishima Y, Hojo H, Shimamura S, Ishikawa F, Tajima S,Tanaka K, Ariyoshi M, Shirakawa M, Ikeguchi M, Kidera A, *Suetake I, *Arita K, *Nakanishi M.
Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance.
Mol. Cell 68,350-360 (2017)
PMID: 29053958 - *Nishiyama A, Mulholland C, Bultmann S, Kori A, Endo A, Saeki Y, Qin W, Trummer C, Chiba Y, Yokoyama H, Kumamoto S, Kawakami T, Hojo H, Nagae G, Aburatani H, Tanaka K, *Arita K, *Leonhardt H, *Nakanishi M.
Two distinct modes of DNMT1 recruitment ensure the stable maintenance DNA methylation.
Nat. Commun. 11, 1222 (2020)
PMID: 32144273