Yusaku Miyamae
Development of a molecular tool for conditional control of cellular protein stability using a excisable degradation domain
 |
Yusaku Miyamae, PhDUniversity of Tsukuba, Faculty of Life and Environmental Sciences |
|---|
Research summary
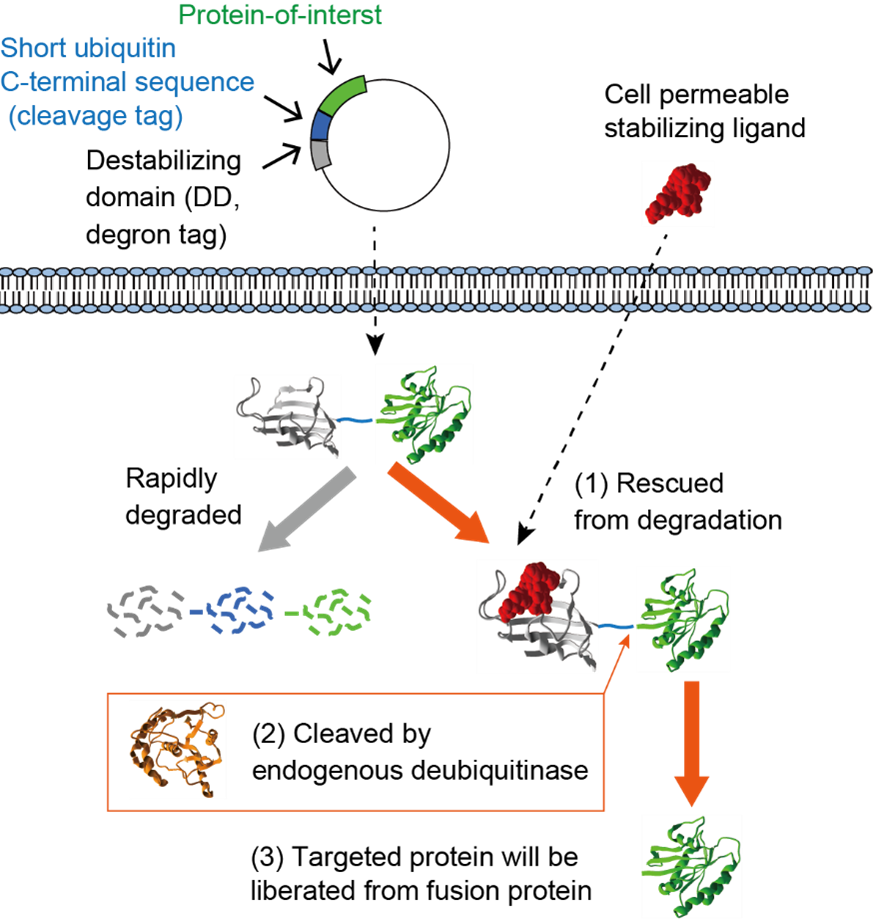
Technology to conditionally regulate protein stability without interfering the function is a powerful tool for biomedical research. Recently, several research groups have developed a variety of methodologies for control of expression level of the target protein using a degron peptide, which confers instability to tagged protein and induce the proteasomal degradation in response to chemical treatment or photo irradiation. Unlike RNAi and Cre/loxP systems, these methods regulate the protein stability in post-translational level and enable to control the protein expression within 30-60 min after the stimulation. However, the most of degron systems require permanent fusion of targeted protein with a degron tag, which may disrupt the molecular function of the target in some cases. Previously we have developed a complementary method to conditionally control cellular protein stability by introducing a ubiquitin variant between destabilizing domain (DD) and targeted protein. In the absence of the stabilizing ligand, the DD dominates and the entire polypeptide is rapidly degraded by the ubiquitin-proteasome system. In the presence of stabilizing ligand, the fusion protein is stabilized and becomes a substrate for abundant ubiquitin-specific proteases (informally called deubiquitinating enzyme or DUB), liberating a native or a near-native protein of interest (POI) from the DD tag. This technique is useful for the regulation of a protein whose free N-terminus is required for its biological function. In this project, we will develop an improved system using short ubiquitin C-terminal sequence to allow more efficient release of a POI from a DD compared to the previous method. The method developed in this project will also be applied for the regulation of endogenous protein expression by combining with genome editing technology.
Former Publications
- Ohtera A, *Miyamae Y, Yoshida K, Maejima K, Akita T, Kakizuka A, Irie K, Masuda S, Kambe T, *Nagao M. Identification of a new type of covalent PPARg agonist using a ligand-linking strategy.
- *Miyamae Y, Nishito Y, Nakai N, Nagumo Y, Usui T, Masuda S, Kambe T, *Nagao M. Tetrandrine induces lipid accumulation through blockade of autophagy in a hepatic stellate cell line.
- Utsugi Y, Kobuchi H, Kawamura Y, Atito ASA, Nagao M, Isoda H, *Miyamae Y. Importance of the proximity and orientation of ligand-linkage to the design of cinnamate-GW9662 hybrid compounds as covalent PPARg agonists.
- Kurata M, Fujiwara N, Fujita K, Yamanaka Y, Seno S, Kobayashi H, Miyamae Y, Takahashi N, Shibuya M, *Masuda S. Food-derived compounds apigenin and luteolin modulate mRNA splicing of introns with weak splice sites.
- Miyamae Y, Chen LC, Utsugi Y, Farrants H, *Wandless T J. A method for conditional regulation of protein stability in native or near-native form.
ACS Chem. Biol. 10, 2794-2804 (2015)
PMID: 26414848
Biochem. Biophys. Res. Commun. 477, 40-46 (2016)
PMID: 27270032
Molecules 24, 2019 (2019)
PMID: 31137814
iScience 22, 336-352 (2019)
PMID: 31809999
Cell Chem. Biol. 27, 1573-1581 (2020)
PMID: 33007216