Yoshitaka Matsuo
Deciphering ribosome-ubiquitin code using chemo-technologies and its control
 |
Yoshitaka Matsuo, PhDInstitute of Medical Science, The University of Tokyo, Division of RNA and Gene regulation |
|---|
Research summary
Over the past decades, translational regulation is mainly controlled by a variety of RNA-binding proteins, translation-associated factors, and numerous enzymes; however, recent studies have been uncovering the expanding roles of ribosome ubiquitination in translational control.
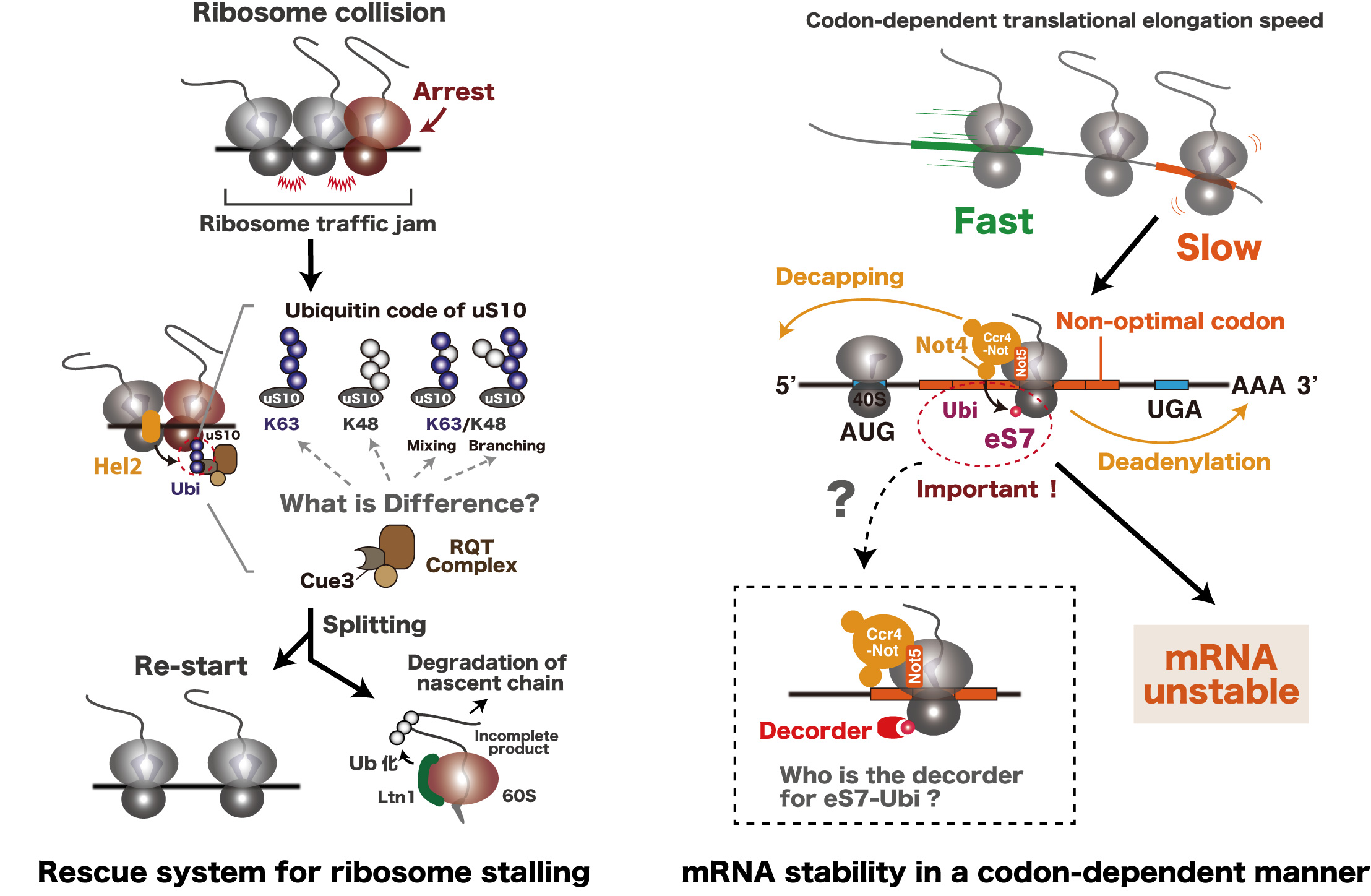
Ribosome-associated quality control (RQC) represents a rescue pathway in eukaryotic cells triggered upon translational stalling. This pathway is triggered by Hel2, which monitors translation and recognizes ribosomes that collide as a consequence of translational stalling. These collided ribosomes are ubiquitinated by Hel2 on ribosomal protein uS10, which marks them as substrates for the RQC pathway (Fig: Left panel). Hence, this ubiquitination triggers ribosome subunit dissociation to allow the downstream RQC processes to be executed.
Control of messenger RNA (mRNA) decay rate is intimately connected to translation elongation, which is monitored by the Ccr4-Not complex. The Ccr4-Not complex preferentially binds to the E-site of translating ribosome at the non-optimal codon via the Not5 N-terminal region (Fig: Right panel). Furthermore, the ubiquitination of eS7 by Not4 is essential for the destabilization of non-optimal mRNAs in this system (Fig: Right panel).
In this way, the ribosome ubiquitination has a critical role in the response to the dynamics of translation elongation; however, the mechanistic details of the recognition and action mode in the ubiquitin chain type-dependent manner are largely unknown. Thus, we here focus on the two types of ribosome ubiquitination: uS10- and eS7-ubiquitin chains and will address the role of them on the RQC and mRNA stabilizing system, respectively. In parallel, we will attempt to develop novel chemical tools, which enable us to control the individual ribosome-ubiquitin code.
Publications
- Li S, Ikeuchi K, Kato M, Buschauer R, Sugiyama T, Adachi S, Kusano H, Natsume T, Berninghausen O, Matsuo Y, Becker T, *Beckmann R, *Inada T.
Sensing of individual stalled 80S ribosomes by Fap1 for non-functional rRNA turnover.
Mol Cell. 82, 3424-3437 (2022)
PMID: 36113412 - Narita M, Denk T, Matsuo Y, Sugiyama T, Kikuguchi C, Ito S, Sato N, Suzuki T, Hashimoto S, Machova I, Tesina P, *Beckmann R, *Inada T.
A distinct human disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation.
Nat Commun. 13, 6411 (2022)
PMID: 36302773 - *Matsuo Y, Uchihashi T, *Inada T.
Decoding of the ubiquitin code for clearance of colliding ribosomes by the RQT complex.
Nat Commun. 14, 79 (2023)
PMID: 36627279 - Tomomatsu S, Watanabe A, Tesina P, Hashimoto S, Ikeuchi K, Li S, Matsuo Y, Beckmann R, *Inada T.
Two modes of Cue2-mediated mRNA cleavage with distinct substrate recognition initiate No-go decay.
Nucleic Acids Res. 51, 253-270 (2023)
PMID: 36583309 - *Tesina P, Ebine S, Buschauer R, Thoms M, Matsuo Y, *Inada T, *Beckmann R.
Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol Cell. 83, 607-621 (2023)
PMID: 36804914 - Best K, Ikeuchi K, Kater L, Best D, Musial J, Matsuo Y, Berninghausen O, Becker T, *Inada T, *Beckmann R.
Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun. 14, 921 (2023)
PMID: 36801861
Former Publications
- Matsuo Y, Granneman S, Thoms M, Manikas RG, Tollervey D, *Hurt E.
- Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K, Ingolia NT, Beckmann R, *Inada T.
- Matsuo Y, Tesina P, Nakajima S, Mizuno M, Endo A, Buschauer R, Cheng J, Shounai O, Ikeuchi K, Saeki Y, Becker T, *Beckmann R, *Inada T.
RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1.
Nat. Struct. Mol. Biol. 27, 323-332 (2020)
PMID: 32203490 - Buschauer R, Matsuo Y, Sugiyama T, Chen YH, Alhusaini N, Sweet T, Ikeuchi K, Cheng J, Matsuki Y, Nobuta R, Gilmozzi A, Berninghausen O, Tesina P, Becker T, *Coller J, *Inada T, *Beckmann R.
- *Matsuo Y ,*Inada T.
Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export.
Nature. 505, 112-116 (2014)
PMID: 24240281
Ubiquitination of stalled ribosome triggers ribosome-associated quality control.
Nat. Commun. 8, 159 (2017)
PMID: 28757607
The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science. 368, eaay6912 (2020)
PMID: 32299921
The ribosome collision sensor Hel2 functions as preventive quality control in the secretory pathway.
Cell Rep. 34, 108877 (2021)
PMID: 33761353