Toshifumi Inada
Molecular basis of RQC control via targeting of specific ubiquitin chains
 |
Toshifumi Inada, PhDInstitute of Medical Science, The University of Tokyo |
|---|
Research summary
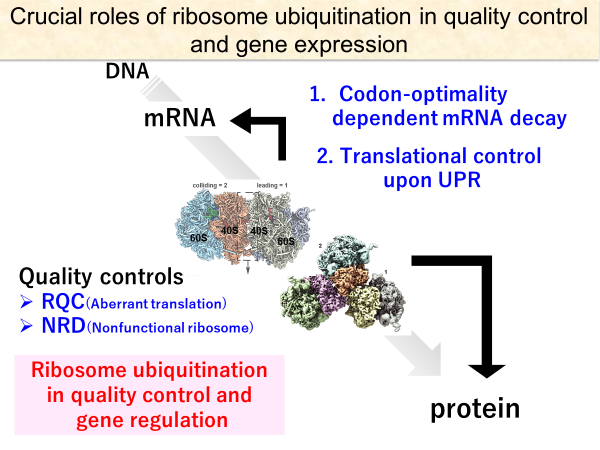
Ribosome-associated quality control (RQC) represents a rescue pathway in eukaryotic cells triggered upon translational stalling. The collided ribosomes consist of the leading stalled ribosome and the following colliding ribosomes. The crucial E3 ligase Hel2 recognizes collided ribosomes and ubiquitinates the specific 40S ribosomal protein uS10. Subsequently, the RQC trigger (RQT) complex dissociates the ubiquitinated ribosomes into the subunits. The distinct collided ribosomes architecture visualized by cryo-EM provides the structural basis for more efficient recognition by Hel2. We reconstituted Hel2-dependent poly-ubiquitination and RQT complex-mediated subunit dissociation reactions in vitro. We found that the RQT complex preferentially dissociates the first stalled poly-ubiquitinated ribosome in an ATP-dependent manner. These findings provide fundamental mechanistic insights into RQC, and its physiological role in maintaining cellular protein homeostasis, including He2-mediated force quit of translation as preventive quality control in the secretory pathway.
We are also investigating the crucial roles of ribosome ubiquitination in post-translational regulations. We recently found that ribosome ubiquitination plays a crucial role in the regulation of mRNA stability, depending on a codon-optimality. The eS7 ubiquitination facilitates the recruitment of the Ccr4-NOT complex to the elongating ribosome, depending on a codon-optimality. The ribosome ubiquitination also plays a crucial role in an eIF2a-phosphorylation independent translational control upon ER stress response. In collaboration with the members of the “chemo-ubiquitin”, we apply novel chemical approaches to understand the molecular mechanism of ribosome ubiquitination-mediated post-translational regulations.
Publications
- Yasuda S, Tsuchiya H, Kaiho A, Guo Q, Ikeuchi K, Endo A, Arai N, Ohtake F, Murata S, Inada T, Baumeister W, Fernandez-Busnadiego R, Tanaka K, *Saeki Y.
Stress- and ubiquitylation-dependent phase separation of the proteasome.
Nature 578, 296-300 (2020)
PMID: 32025036
Highlighted in Cell Res.
EurekAlert!
Faculty Opinions - Matsuo Y, Tesina P, Nakajima S, Mizuno M, Endo A, Buschauer R, Cheng J, Shounai O, Ikeuchi K, Iwasaki S, Saeki Y, Becker T, *Beckmann R, *Inada T.
RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1.
Nat. Stuct. Mol. Biol. 27, 323-332 (2020)
PMID: 32203490 -
Buschauer R, Matsuo Y (co-1st author), Sugiyama T, Chen YH, Alhusaini N, Sweet T, Ikeuchi K, Cheng J, Matsuki Y, Gilmozzi A, Berninghausen O, Becker T, *Coller J, *Inada T, *Beckmann R.
The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science 368, 6448 (2020)
PMID: 32299921 - Hashimoto S, Sugiyama T, Yamazaki R, Nobuta R,*Inada T.
Identification of a novel trigger complex that facilitates ribosome-associated quality control in mammalian cells.
Sci. Rep. 10, 3422(2020)
PMID: 32099016 - Matsuki Y, Matsuo Y, Nakano Y, Iwasaki S, Yoko H, Udagawa T, Li S, Saeki Y, Yoshihisa T, Tanaka K, Ingolia N, *Inada T.
Ribosomal protein S7 ubiquitination during ER stress in yeast is associated with selective mRNA translation and stress outcome.
Sci. Rep. 10, 19669 (2020)
PMID: 33184379 - Udagawa T Seki M, Okuyama T, Adachi S, Natsume T, Noguchi T, Matsuzawa A, *Inada T.
Failure to degrade CAT-tailed proteins disrupts neuronal morphogenesis and cell survival.
Cell Rep. 34, 108599(2021)
PMID: 33406423
Former Publications
- Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, *Inada T.
Dom34:Hbs1 Plays a General Role in Quality-Control Systems by Dissociation of a Stalled Ribosome at the 3′ End of Aberrant mRNA.
Mol. Cell 46, 518-529 (2012)
PMID: 22503425 - Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K, Ingolia NT, Beckmann R, *Inada T.
Ubiquitination of Stalled Ribosome Triggers Ribosome-associated Quality Control.
Nat. Commun. 8, 159 (2017)
PMID: 28757607 - *Inada T.
The ribosome as a platform for mRNA and nascent polypeptide quality control.
Trends Biochem. Sci. 42, 5-15 (2017)
PMID: 27746049 - Ikeuchi K, Tesina P, Matsuo Y, Sugiyama T, Cheng J, Saeki Y, Tanaka K, Becker T, *Beckmann R, *Inada T.
Collided ribosomes form a unique structural interface to induce Hel2‐driven quality control pathways.
EMBO J. 38. e100276 (2019)
PMID: 30609991 - Sugiyama T, Li S, Kato M, Ikeuchi K, Ichimura A, Matsuo Y, *Inada T.
Sequential ubiquitination of ribosomal protein uS3 triggers the degradation of non-functional 18S rRNA.
Cell Rep. 26, 3400-3415.e7 (2019)
PMID: 30893611