Shigetsugu Hatakeyama
Physicochemical properties and molecular regulation of TRIM-type ubiquitin ligases
 |
Shigetsugu Hatakeyama, MD, PhDDepartment of Biochemistry, Faculty of Medicine, Hokkaido University |
|---|
Research summary
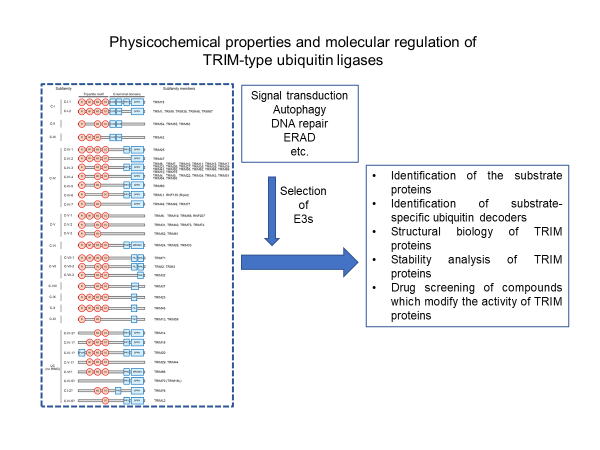
TRIM family is a family of RING-type E3 ubiquitin ligases (about 70 in humans) and has been reported to be involved in various functions including the regulation of cancer, immunity, development, autophagy. The purpose of this study is to analyze the physicochemical properties of TRIM proteins as "proteins" to examine their relevance to function and to accumulate knowledge in drug discovery research in the future.
In order to elucidate the function of TRIM protein, it is first necessary to identify its substrate proteins and to determine the structure of TRIM proteins. It is important to determine the targeted TRIM protein based on biological and clinical information and the relationship with the disease, and to conduct compound screening that results in their functional changes. To that end, we will establish a method for identifying highly specific substrate proteins of TRIM proteins and ubiquitin decoders (TUBE-E3 method). At the same time, it is necessary to proceed with structural biological analysis of TRIM proteins. The E3 candidates analyzed in this project will be mainly TRIM proteins that are thought to be involved in signal transduction, selective autophagy, DNA repair and ERAD.
From the viewpoints of proteomics, structural biology, and chemical biology, the following methods will be used to accumulate basic knowledge useful for "the development of chemical tools" and "drug discovery" by promoting the analysis of target E3.
Publications
- Takahashi H, Ranjan A, Chen S, Suzuki H, Shibata M, Hirose T, Hirose H, Sasaki K, Abe R, Chen K, He Y, Zhang Y, Takigawa I, Tsukiyama T, Watanabe M, Fujii S, Iida M, Yamamoto J, Yamaguchi Y, Suzuki Y, Matsumoto M, Nakayama KI, Washburn M, Saraf A, Florens L, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, *Hatakeyama S.
The role of Mediator and Little Elongation Complex in transcription termination.
Nature Commun. 11, 1063 (2020)
PMID: 32102997 - Sugawara E, *Kato M, Kudo Y, Lee W, Hisada R, Fujieda Y, Oku K, Bohgaki T, Amengual O, Yasuda S, Onodera T, Hatakeyama S, Atsumi T.
Autophagy promotes citrullination of VIM (vimentin) and its interaction with major histocompatibility complex class II in synovial fibroblasts.
Autophagy 16, 946-955(2020)
PMID: 31486697 - Sugisawa E, Takayama Y, Takemura N, Kondo T, Hatakeyama S, Kumagai Y, Sunagawa M, Tominaga M, *Maruyama K.
RNA sensing by gut Piezo1 is essential for systematic serotonin synthesis.
Cell 182, 609-624 (2020)
PMID: 32640190
Former Publications
- Masuda Y, Takahashi H, Sato S, Tomomori-Sato C, Saraf A, Washburn WP, Florens L, Conaway RC, Conaway JW,*Hatakeyama S.
TRIM29 regulates the assembly of DNA repair proteins into damaged chromatin.
Nat. Commun. 6, 7299 (2015)
PMID: 26095369 - Mizushima W, Takahashi H, Watanabe M, Kinugawa S, Matsushima S, Takada S, Yokota T, Furihata T, Matsumoto J, Tsuda M, Chiba I, Nagashima S, Yanagi S, Matsumoto M, Nakayama KI, Tsutsui H, *Hatakeyama S.
The novel heart-specific RING finger protein 207 is involved in energy metabolism in cardiomyocytes.
J. Mol. Cell. Cardiol. 100, 43-53 (2016)
PMID: 27677939 - *Hatakeyama S.
TRIM Family Proteins: Roles in Autophagy, Immunity and Carcinogenesis.
Trends Biochem. Sci. 42, 297-311 (2017)
PMID: 28118948 - Sang Y, Li Y, Song L, Alvarez AA, Zhang W, Lv D, Tang J, Liu F, Chang Z, Hatakeyama S, Hu B, Cheng S,*Feng H.
TRIM59 promotes gliomagenesis by inhibiting TC45 dephosphorylation of STAT3.
Cancer Res. 78, 1792-1804 (2018)
PMID: 29386185 - *Yanagi T, Watanabe M, Hata M, Kitamura S, Imafuku K, Yanagi H, Homma MD A, Wang L, Takahashi H, Shimizu H, *Hatakeyama S.
Loss of TRIM29 alters keratin distribution to promote cell invasion in squamous cell carcinoma.
Cancer Res. 78, 6795-6806 (2018)
PMID: 30389700