Research Group
TeamResearch Group
Research group A02: Development of ubiquitin chemo-technologies
Chemical biology for controlling ubiquitin function
Minoru Yoshida, PhDRIKEN Center for Sustainable Resource Science (CSRS) |
Protein ubiquitination has been shown to function for not only proteolytic degradation by the proteasome but also a variety of protein functional alterations, which exceed our initial assumptions. The basis for this diverse function of ubiquitination is the diversity of molecules (decoders) that interact with the structurally different linkages of ubiquitin modification. In this research (Scientific Research on Innovative Areas “Ubiquitin new frontier driven by Chemo-technologies”), we are trying to develop a new methodology to understand and regulate ubiquitin-modification systems by making full use of a chemical approach (chemotechnology). In collaboration with other members in the research group, we will develop two types of chemical tools: a new ubiquitin-targeting chimeric compounds named UbTACs and specific inhibitors for the interaction between ubiquitin chains and their decoders. UbTACs, which are developed from the basic concept of proteolysis targeting chimeras (PROTACs), are expected to introduce ubiquitin modification into particular target proteins, which may result in endocytosis and selective autophagy as well as target protein knockdown. Ubiquitin-decoder interaction inhibitors will be useful for both basic understanding of the physiological roles of specific ubiquitin modifications and novel drug discovery. We will also serve as a hub for collaboration between members in biological and chemical fields in the research group.
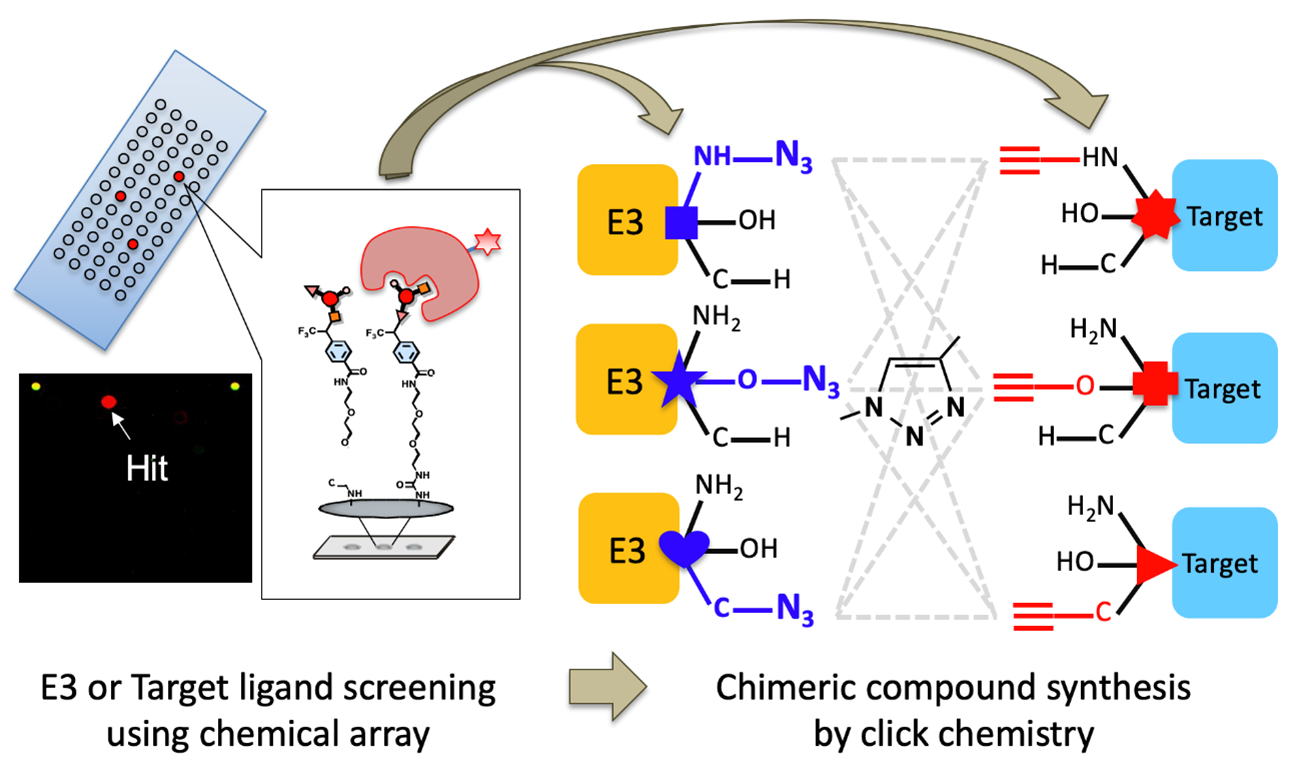
Strategy for development of novel ubiquitin-targeting chimeric compounds named UbTACs.
Chemical biology for controlling ubiquitin function: Ligand discovery for development of UbTACs
Makoto Kawatani, PhDBiomolecular Characterization Unit, RIKEN Center for Sustainable Resource Science (CSRS) Senior Research Scientist |
|
Yasumitsu Kondoh, PhD(Until 2021)RIKEN Center for Sustainable Resource Science, Chemical Biology Research Group |
We are attempting to pioneer a new methodology to understand and operate ubiquitin-modified systems using a chemical approach (chemotechnology). This research aims to develop the tools that serve as the foundation for new methodology. The key for this research is new ubiquitination-inducing chimeric molecules, “Ubiquitin-TArgeting Chimeras (UbTACs)”. Chimeric compounds called SNIPER and PROTACs ubiquitinate non-original target substrate proteins (neosubstrates) by hijacking ubiquitin ligase, followed by inducing degradation. By developing this technology, we aim to create new UbTACs molecules that can induce not only knockdown of neosubstrates but also endocytosis, signal transduction, and autophagy. In order to achieve these, it is necessary to explore new E3 ligands and neosubstrate ligands, and also to develop a technique to chimerize them. We have original chemical array technology, which enables ultra high-throughput screening of ligands for proteins of interest. The chemical array has approximately 22,000 library compounds from the RIKEN Natural Products Depository (NPDepo) immobilized on glass slides by a photo-crosslinking method. Using chemical array, we will search for novel E3 ligands and neosubstrate ligands. As second screening, the hit compounds of the chemical array screening will be analyzed using two-dimensional SPR or Biacore and candidates will be narrowed down based on binding affinity (Fig. 1). Finally, we are going to identify functional groups of explored candidate compounds available for chimerization through structure-activity relationship studies.
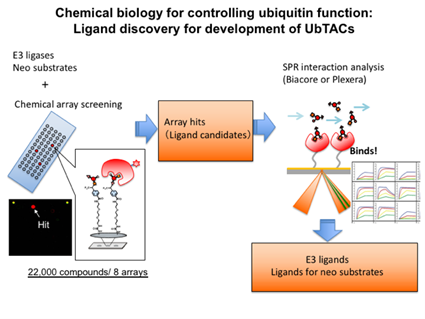
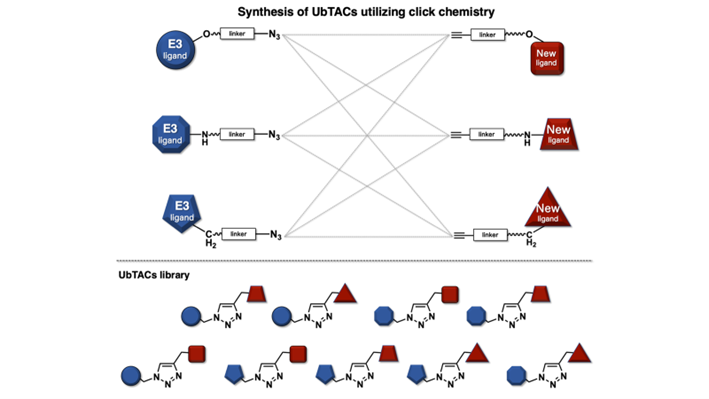
Chemical biology for controlling ubiquitin function: Development of UbTACs by utilizing click chemistry
Ambara Rachmat PradiptaTokyo Institute of Technology, School of Materials and Chemical Technology, |
Ubiquitin-targeting chimeras (UbTACs) is an approach to catalyze the ubiquitination of substrate proteins to trigger various cellular outcomes. We aim to design the UbTACs that comprise of a novel E3 ubiquitin ligase ligand conjugated via a linker to a neo-substrate protein-ligand. To develop potent UbTACs, we need to examine the structure-activity relationship of these bispecific molecules. Herein, the click reaction will be utilized to couple the novel E3 ubiquitin ligase ligands and neo-substrate protein ligands. The click reaction of azides and alkynes to form a 1,2,3-triazole ring, which arguably is one of the most reliable bonds forming reaction, is easy to use and can be performed under mild reaction condition. We will prepare the azide-linker and alkyne-linker derivatives, which would allow for the parallel synthesis of the UbTACs library.
Publications
- Sohtome Y, Shimazu T, Barjau J, Fujishiro S, Akakabe M, Terayama N, Dodo K, Ito A, Yoshida M, Shinkai Y, Sodeoka M.
Unveiling epidithiodiketopiperazine as a non-histone arginine methyltransferase inhibitor by chemical protein methylome analyses.
Chem. Commun. 54, 9202-9205 (2018)
PMID: 30063235 - Hiranaka S, Tega Y, Higuchi K, Kurosawa T, Deguchi Y, Arata M, Ito A, Yoshida M, Nagaoka Y, Sumiyoshi T.
Design, Synthesis, and Blood-Brain Barrier Transport Study of Pyrilamine Derivatives as Histone Deacetylase Inhibitors.
ACS Med. Chem. Lett. 9, 884-888 (2018)
PMID: 30258535 - Furuta H, Yoshihara H, Fukushima T, Yoneyama Y, Ito A, Worrall C, Girnita A, Girnita L, Yoshida M, Asano T, Komada M, Kataoka N, Chida K, Hakuno F, Takahashi SI.
IRS-2 deubiquitination by USP9X maintains anchorage-independent cell growth via Erk1/2 activation in prostate carcinoma cell line.
Oncotarget 9, 33871-33883 (2018)
PMID: 30338032 - Nakamoto Y, Pradipta AR, Mukai H, Zouda M, Watanabe Y, Kurbangalieva A, Ahmadi P, Manabe Y, Fukase K, Tanaka K.
Expanding the applicability of the metal labeling of biomolecules by the RIKEN click reaction: A case study with Gallium-68 positron emission tomography.
ChemBioChem 19, 2055-2060 (2018)
PMID: 30066425 - Simpkins SW, Nelson J, Deshpande R, Li SC, Piotrowski JS, Wilson EH, Gebre AA, Safizadeh H, Okamoto R, Yoshimura M, Costanzo M, Yashiroda Y, Ohya Y, Osada H, Yoshida M, Boone C, Myers CL.
Predicting bioprocess targets of chemical compounds through integration of chemical-genetic and genetic interactions.
PLoS Comput. Biol. 14, e1006532 (2018)
PMID: 30376562 - Tanei T, Pradipta AR, Morimoto K, Fujii M, Arata M, Ito A, Yoshida M, Saigitbatalova E, Kurbangalieva A, Ikeda JI, Morii E, Noguchi S, Tanaka K.
Cascade reaction in human live tissue allows clinically applicable diagnosis of breast cancer morphology.
Adv. Sci. 6, 1801479 (2018)
PMID: 30693189 - Mizutani A, Yashiroda Y, Muramatsu Y, Yoshida H, Chikada T, Tsumura T, Okue M, Shirai F, Fukami T, Yoshida M, Seimiya H.
RK-287107, a potent and specific tankyrase inhibitor, blocks colorectal cancer cell growth in a preclinical model.
Cancer Sci. 109, 4003-4014 (2018)
PMID: 30238564 - Simpkins SW, Deshpande R, Nelson J, Li SC, Piotrowski JS, Ward HN, Yashiroda Y, Osada H, Yoshida M, Boone C, Myers CL.
Using BEAN-counter to quantify genetic interactions from multiplexed barcode sequencing experiments.
Nat. Protoc. 14, 415-440 (2019)
PMID: 30635653 - Suvarna K, Honda K, Muroi M, Kondoh Y, Osada H, *Watanabe N.
A small-molecule ligand of valosin-containing protein/p97 inhibits cancer cell-accelerated fibroblast migration.
J. Biol. Chem. 294, 2988-2996 (2019)
PMID: 30610116 - Ling F, Bradshaw E, Yoshida M.
Prevention of mitochondrial genomic instability in yeast by the mitochondrial recombinase Mhr1.
Sci. Rep. 9, 5433 (2019)
PMID: 30931958 - Shirai F, Tsumura T, Yashiroda Y, Yuki H, Niwa H, Sato S, Chikada T, Koda Y, Washizuka K, Yoshimoto N, Abe M, Onuki T, Mazaki Y, Hirama C, Fukami T, Watanabe H, Honma T, Umehara T, Shirouzu M, Okue M, Kano Y, Watanabe T, Kitamura K, Shitara E, Muramatsu Y, Yoshida H, Mizutani A, Seimiya H, Yoshida M, Koyama H.
Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem. 62, 3407-3427 (2019)
PMID: 30883102 - Shu K, Iwamoto N, Honda K, Kondoh Y, Hirano H, Osada H, Ohno H, Fujii N, Oishi S.
Development of mirror-image screening systems for XIAP BIR3 domain inhibitors.
Bioconjug. Chem. 30, 1395-1404 (2019)
PMID: 30888797 - Qi Y, Zhao X, Chen J, Pradipta AR, Wei J, Ruan H, Zhou L, Hsung RP, Tanaka K.
In vitro and in vivo cancer cell apoptosis triggered by competitive binding of Cinchona alkaloids to the RING domain of TRAF6.
Biosci. Biotechnol. Biochem. 83, 1011-1026 (2019)
PMID: 31074699 - Pradipta AR, Fujii M, Tanei T, Morimoto K, Shimazu K, Noguchi S, *Tanaka K.
Tetramethylrhodamine is an essential scaffold of azide probe in detecting cellular acrolein.
Bioorg. Med. Chem. 27, 2228-2234 (2019)
PMID: 31023557 - Murashima A, Shinjo K, Katsushima K, Onuki T, Kondoh Y, Osada H, Kagaya N, Shin-Ya K, Kimura H, Yoshida M, Murakami S, Kondo Y.
Identification of a chemical modulator of EZH2-mediated silencing by cell-based high-throughput screening assay.
J. Biochem. 166, 41-50 (2019)
PMID: 30690451 - Mitsui E, Yoshida S, Shinoda Y, Matsumori Y, Tsujii H, Tsuchida M, Wada S, Hasegawa M, Ito A, Mino K, Onuki T, Yoshida M, Sasaki R, Mizukami T.
Identification of ryuvidine as a KDM5A inhibitor.
Sci. Rep. 9, 9952 (2019)
PMID: 31289306 - Takase S, Kurokawa R, Kondoh Y, Honda K, Suzuki T, Kawahara T, Ikeda H, Dohmae N, Osada H, Shin-Ya K, Kushiro T, *Yoshida M, *Matsumoto K.
Mechanism of action of prethioviridamide, an anticancer ribosomally synthesized and post-translationally modified peptide with a polythioamide structure.
ACS Chem. Biol. 14, 1819-1828 (2019)
PMID: 31365229 - Yoshida K, Kondoh Y, Iwahashi F, Nakano T, Honda K, Nagano E, Osada H.
Abscisic acid derivatives with different alkyl chain lengths activate distinct abscisic acid receptor subfamilies.
ACS Chem. Biol. 14, 1964-1971 (2019)
PMID: 31497942 - Lodochnikova OA, Chulakova DR, Gerasimova DP, Litvinov IA, Pradipta AR, *Tanaka K, *Kurbangalieva AR.
Stereochemical features of the crystallization of eight-membered 1,5-diazaheterocycles with chiral aminoindanol fragments at nitrogen atoms.
J. Struct. Chem. 61, 119-125 (2020) - Chulakova DR, *Pradipta AR, Lodochnikova OA, Kuznetsov DR, Bulygina KS, Smirnov IS, Usachev KS, Latypova LZ, Kurbangalieva AR, *Tanaka K.
Facile access to optically active 2,6-dialkyl-1,5-diazacyclooctanes.
Chem. Asian J. 14, 4048-4054 (2019)
PMID: 31381243 - Suvarna K, Honda K, Muroi M, Kondoh Y, Osada H, *Watanabe N.
Measurement of ATPase activity of valosin-containing protein/p97.
Bio-protocol 10, e3516 (2020) - Suvarna K, Honda K, Muroi M, Kondoh Y, *Watanabe N, *Osada H.
Identification of target protein for bio-active small molecule using photo-cross linked beads and MALDI-TOF mass spectrometry.
Bio-protocol 10, e3517 (2020) - *Pradipta AR, Tanei T, Morimoto K, Shimazu K, Noguchi S, *Tanaka K.
Emerging technologies for real-time intraoperative margin assessment in future breast-conserving surgery.
Adv. Sci. 7, 1901519 (2020)
PMID: 32382473 - Han P, Shichino Y, Schneider-Poetsch T, Mito M, Hashimoto S, Udagawa T, Kohno K, Yoshida M, Mishima Y, Inada T, *Iwasaki S.
Genome-wide survey of ribosome collision.
Cell Rep. 31,107610 (2020)
PMID: 32375038 - Jo T, *Nishikori M, Kogure Y, Arima H, Sasaki K, Sasaki Y, Nakagawa T, Iwai F, Momose S, Shiraishi A, Kiyonari H, Kagaya N, Onuki T, Shin-Ya K, Yoshida M, Kataoka K, Ogawa S, *Iwai K, Takaori-Kondo A.
LUBAC accelerates B-cell lymphomagenesis by conferring resistance to genotoxic stress on B cells.
Blood 136, 84-697 (2020)
PMID: 32325488 - Sako K, Futamura Y, Shimizu T, Matsui A, Hirano H, Kondoh Y, Muroi M, Aono H, Tanaka M, Honda K, Shimizu K, Kawatani M, Nakano T, Osada H, Noguchi K, *Seki M.
Inhibition of mitochondrial complex I by the novel compound FSL0260 enhances high salinity-stress tolerance in Arabidopsis thaliana.
Sci. Rep. 10, 8691 (2020)
PMID: 32457324 - *Pradipta AR, Chang TC, *Tanaka K.
Enantioselective synthesis of cyclic and linear diamines by iminocycloaddition.
Chirality 32, 1160-1168 (2020)
PMID: 32621328 - Ikeda H, Muroi M, Kondoh Y, Ishikawa S, Kakeya H, Osada H, *Imoto M.
Miclxin, a novel MIC60 inhibitor, induces apoptosis via mitochondrial stress in β-catenin mutant tumor cells.
ACS Chem. Biol. 15, 2195-2204 (2020)
PMID: 32584541 - Kawamura T, Futamura Y, Shang E, Muroi M, Janning P, Ueno M, Wilke J, Takeda S, Kondoh Y, Ziegler S, Watanabe N, Waldmann H, *Osada H.
Discovery of small-molecule modulator of heterotrimeric Gi-protein by integrated phenotypic profiling and chemical proteomics.
Biosci. Biotechnol. Biochem. 84, 2484-2490 (2020)
PMID: 32867616 - Radwan MO, Ciftci HI, Ali TFS, Koga R, Tateishi H, Nakata A, Ito A, Yoshida M, Fujita M, *Otsuka M.
Structure activity study of S-trityl-cysteamine dimethylaminopyridine derivatives as SIRT2 inhibitors: Improvement of SIRT2 binding and inhibition.
Bioorg. Med. Chem. Lett. 30,127458 (2020)
PMID: 32755678 - Kobayashi H, Nishimura H, Kudo N, Osada H, *Yoshida M.
A novel GSK3 inhibitor that promotes self-renewal in mouse embryonic stem cells.
Biosci. Biotechnol. Biochem. 84, 2113-2120 (2020)
PMID: 32640867 - Fujita K, Kondoh Y, Honda K, Haga Y, Osada H, Matsumura C, *Inui H.
Pesticide treatment reduces hydrophobic pollutant contamination in Cucurbita pepo through competitive binding to major latex-like proteins.
Environ. Pollut. 266, 115179 (2020).
PMID: 32717636 - Sophonnithiprasert T, Aruksakunwong O, Tashiro E, Kondoh Y, Muroi M, Osada H, Imoto M, Watanapokasin R.
Interaction between goniothalamin and peroxisomal multifunctional enzyme type 2 triggering endoplasmic reticulum stress.
Heliyon 6, e05200 (2020)
PMID: 33102840 - Smirnov I, Sibgatullina R, Urano S, Tahara T, Ahmadi P, Watanabe Y, Pradipta AR, Kurbangalieva A, *Tanaka K.
A Strategy for Tumor Targeting by Higher-Order Glycan Pattern Recognition: Synthesis and In Vitro and In Vivo Properties of Glycoalbumins Conjugated with Four Different N-Glycan Molecules.
Small 16, 2004831 (2020)
PMID: 33079456 - Muguruma K, Pradipta AR, Ode Y, Terashima K, Michiba H, Fujii M, *Tanaka K.
Disease-associated acrolein: A possible diagnostic and therapeutic substrate for in vivo synthetic chemistry.
Bioorg. Med. Chem. 28, 115831 (2020)
PMID: 33199202 - Kobayashi H, Hatakeyama H, Nishimura H, Yokota M, Suzuki S, Tomabechi Y, Shirouzu M, Osada H, Mimaki M, *Goto Y, *Yoshida M.
Chemical reversal of abnormalities in cells carrying mitochondrial DNA mutations.
Nat. Chem. Biol. 171,335-343 (2021)
PMID: 33168978 - Yoshimoto R, Chhipi-Shrestha JK, Schneider-Poetsch T, Furuno M, Maxwell Burroughs A, Noma S, Suzuki H, Hayashizaki Y, Mayeda A, Nakagawa S, Kaida D, Iwasaki S, *Yoshida M.
Spliceostatin A interaction with SF3B1 limits U1 snRNP availability and causes premature cleavage and polyadenylation.
Cell Chem. Biol. 28, 1356-1365 (2021)
PMID: 33784500 - *Pradipta A R, Ahmadi P, Terashima K, Muguruma K, Fujii M, Ichino T, Maeda S, *Tanaka K.
Targeted 1,3-dipolar cycloaddition with acrolein for cancer prodrug activation.
Chem. Sci. 12, 5438-5449 (2021) - Pradipta A R, *Tanaka K.
Application of acrolein imines to organic synthesis, biofunctional studies, and clinical practice.
Chem. Rec. 21, 646-662 (2021)
PMID: 33769681 - Nagashima S, Maruyama J, Honda K, Kondoh Y, Osada H, Nawa M, Nakahama KI, Ishigami-Yuasa M, Kagechika H, Sugimura H, Iwasa H, Arimoto-Matsuzaki K, Nishina H, Hata Y.
CSE1L promotes nuclear accumulation of transcriptional coactivator TAZ and enhances invasiveness of human cancer cells.
J Biol. Chem. 297, 100803 (2021)
PMID: 34022224 - Takayama KI, Honma T, Suzuki T, Kondoh Y, Osada H, Suzuki Y, Yoshida M, Inoue S.
Targeting Epigenetic and Posttranscriptional Gene Regulation by PSF Impairs Hormone Therapy-Refractory Cancer Growth.
Cancer Res. 81, 3495-3508 (2021)
PMID: 33975881 - Yoshida K, Kondoh Y, Nakano T, Bolortuya B, Kawabata S, Iwahashi F, Nagano E, Osada H.
New abscisic acid derivatives revealed adequate regulation of stomatal, transcriptional, and developmental responses to conquer drought.
ACS Chem. Biol. 16, 1566-1575 (2021)
PMID: 34379974 - *Pradipta A R, *Tanaka K.
Biofunctional chemistry and reactivity of biogenic acrolein for cancer diagnosis and therapy.
Chem. Commun. 57, 9798-9806 (2021)
PMID: 34581321 - Nakatani K, Maehama T, Nishio M, Otani J, Yamaguchi K, Fukumoto M, Hikasa H, Hagiwara S, Nishina H, Mak TW, Honma T, Kondoh Y, Osada H, Yoshida M, Suzuki A.
Alantolactone is a natural product that potently inhibits YAP1/TAZ through promotion of reactive oxygen species accumulation.
Cancer Sci. 112, 4303-4316 (2021)
PMID: 34289205 - Sakamoto T, Fukui Y, Kondoh Y, Honda K, Shimizu T, Hara T, Hayashi T, Saitoh Y, Murakami Y, Inoue J, Kaneko S, Osada H, Seiki M.
Pharmacological inhibition of Mint3 attenuates tumour growth, metastasis, and endotoxic shock.
Commun. Biol. 4, 1165 (2021)
PMID: 34621018 - Moon JY, Adams E, Miyazaki T, Kondoh Y, Muroi M, Watanabe N, Osada H, Shin R.
Cesium tolerance is enhanced by a chemical which binds to BETA- GLUCOSIDASE 23 in Arabidopsis thaliana.
Sci. Rep. 11, 21109 (2021)
PMID: 34702872 - Yoshida J, Ohishi T, Abe H, Ohba S, Inoue H, Usami I, Amemiya M, Oriez R, Sakashita C, Dan S, Sugawara M, Kawaguchi T, Ueno J, Asano Y, Ikeda A, Takamatsu M, Amori G, Kondoh Y, Honda K, Osada H, Noda T, Watanabe T, Shimizu T, Shibasaki M, Kawada M.
Mitochondrial complex I inhibitors suppress tumor growth through concomitant acidification of the intra- and extracellular environment.
iScience 24, 103497 (2021)
PMID: 34934919 - Yoshioka H, Kawamura T, Muroi M, Kondoh Y, Honda K, Kawatani M, Aono H, Waldmann H, Watanabe N, Osada H.
Identification of a small molecule that enhances ferroptosis via inhibition of FSP1.
ACS Chem. Biol. 17, 483-491 (2022)
PMID: 35128925 - *Pradipta A R, Michiba H, Kubo A, Fujii M, Tanei T, Morimoto K, Shimazu K, *Tanaka K.
The secon-generation click-to-sense probe for intraoperative diagnosis of breast cancer tissues based on acrolein targeting.
Bull. Chem. Soc. Jpn. (2022) - Chhipi-Shrestha JK, Schneider-Poetsch T, Suzuki T, Mito M, Khan K, Dohmae N, *Iwasaki S, *Yoshida M.
Splicing modulators elicit global translational repression by condensate-prone proteins translated from introns.
Cell Chem. Biol. 29, 259-275 (2022)
PMID: 34520743 - Sasaki K, Suzuki M, Sonoda T, Schneider-Poetsch T, Ito A, Takagi M, Fujishiro S, Sohtome Y, Dodo K, Umehara T, Aburatani H, Shin-ya K, Nakao Y, Sodeoka M, *Yoshida M.
Visualization of the dynamic interaction between nucleosomal histone H3K9 tri-methylation and HP1α chromodomain in living cells.
Cell Chem. Biol. 29, 1153-1161 (2022)
PMID: 35728598 - Kato H, Nemoto K, Shimizu M, Abe A, Asai S, Ishihama N, Matsuoka S, Daimon T, Ojika M, Kawakita K, Onai K, Shirasu K, Yoshida M, Ishiura M, Takemoto D, Takano Y, Terauchi R.
Recognition of pathogen-derived sphingolipids in Arabidopsis.
Science. 376, 857-860 (2022)
PMID: 35587979 - Chhipi-Shrestha JK, Yoshida M, Iwasaki S.
Filter trapping protocol to detect aggregated proteins in human cell lines.
STAR Protoc. 3, 101571 (2022)
PMID: 35880124 - Manzoor M, Muroi M, Ogawa N, Kobayashi H, Nishimura H, Chen D, Fasina OB, Wang J, Osada H, Yoshida M, Xiang L, Qi J.
Isoquercitrin from Apocynum venetum L. produces an anti-obesity effect on obese mice by targeting C-1-tetrahydrofolate synthase, carbonyl reductase, and glutathione S-transferase P and modification of the AMPK/SREBP-1c/FAS/CD36 signaling pathway in mice in vivo.
Food Funct. 13, 10923-10936 (2022)
PMID: 36205648
Former Publications
- Nishimura S, Arita Y, Honda M, Iwamoto K, Matsuyama A, Shirai A, Kawasaki H, Kakeya H, Kobayashi T, Matsunaga S, *Yoshida M.
Marine antifungal theonellamides targets 3ß-hydroxysterol to activate Rho1 signaling.
Nat. Chem. Biol. 6, 519-526 (2010)
PMID: 20543850 - Hirohama M, Kumar A, Fukuda I, Matsuoka S, Igarashi Y, Saitoh H, Takagi M, Shin-ya K, Honda K, Kondoh Y, Saito T, Nakao Y, Osada H, Zhan KY, Yoshida M, *Ito A.
Spectomycin B1 as a novel SUMOylation inhibitor that directly binds to SUMO E2.
ACS Chem. Biol. 8, 2635-2642 (2013)
PMID: 24143955 - Arita Y, Nishimura S, Ishitsuka R, Kishimoto T, Ikenouchi J, Ishii K, Umeda M, Matsunaga S, Kobayashi T, *Yoshida M.
Targeting cholesterol in a liquid-disordered environment by theonellamides modulates cell membrane order and cell shape.
Chem. Biol. 22, 604-610 (2015)
PMID: 25960262 - Ito A, Shimazu T, Maeda S, Shah AA, Tsunoda T, Iemura S-I, Natsume T, Suzuki T, Motohashi H, Yamamoto M, *Yoshida M.
The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1.
Sci. Signal. 8, ra120 (2015)
PMID: 26602019 - Kondoh Y, Honda K, Hiranuma S, Hayashi T, Shimizu T, Watanabe N, *Osada H.
Comparative chemical array screening for p38γ/δ MAPK inhibitors using a single gatekeeper residue difference between p38α/β and p38γ/δ.
Sci. Rep. 6, 29881 (2016)
PMID: 27431267 - Sun X, Hirai G, Ueki M, Hirota H, Wang Q, Hongo Y, Nakamura T, Hitora Y, Takahashi H, Sodeoka M, Osada H, Hamamoto M, *Yoshida M, *Yashiroda Y.
Identification of novel secreted fatty acids that regulate nitrogen catabolite repression in fission yeast.
Sci. Rep. 6, 20856 (2016)
PMID: 26892493 - Nakaoka S, Sasaki K, Ito A, Nakao Y, *Yoshida M.
A genetically encoded FRET probe to detect intranucleosomal histone H3K9 or H3K14 acetylation using BRD4, a BET family member.
ACS Chem. Biol. 11, 729-733 (2016)
PMID: 25946208 - Takase S, Kurokawa R, Arai D, Kanemoto Kanto K, Okino T, Nakao Y, Kushiro T, *Yoshida M, *Matsumoto K.
A quantitative shRNA screen identifies ATP1A1 as a gene that regulates cytotoxicity by aurilide B.
Sci. Rep. 7, 2002 (2017)
PMID: 28515454 - Yoshimoto R, Kaida D, Furuno M, Burroughs AM, Noma S, Suzuki H, Kawamura Y, Hayashizaki Y, Mayeda A, *Yoshida M.
Global analysis of pre-mRNA subcellular localization following splicing inhibition by spliceostatin A.
RNA 23, 47-57 (2017)
PMID: 27754875 - Piotrowski JS, Li SC, Deshpande R, Simpkins SW, Nelson J, Yashiroda Y, Barber JM, Safizadeh H, Wilson E, Okada H, Gebre AA, Kubo K, Torres N, Leblanc MB, Andrusiak K, Okamoto R, Yoshimura M, van Leeuwen J, Shirahige K, Baryshnikova A, Brown G, Saito T, Costanzo M, Andrews B, Ohya Y, *Osada H, *Yoshida M, *Myers CL, *Boone C.
Functional annotation of chemical libraries across diverse biological processes.
Nat. Chem. Biol. 13, 982-993 (2017)
PMID: 28759014