Research Group
Research Group
Research group A01: Elucidation of ubiquitin functions by chemo-technologies
Deciphering the ubiquitin code by mass spectrometry and chemo-technologies
Yasushi Saeki, PhDTokyo Metropolitan Institute of Medical Science, Protein Metabolism Project |
|
Fumiaki Ohtake, PhDHoshi University, Institute for Advanced Life Sciences |
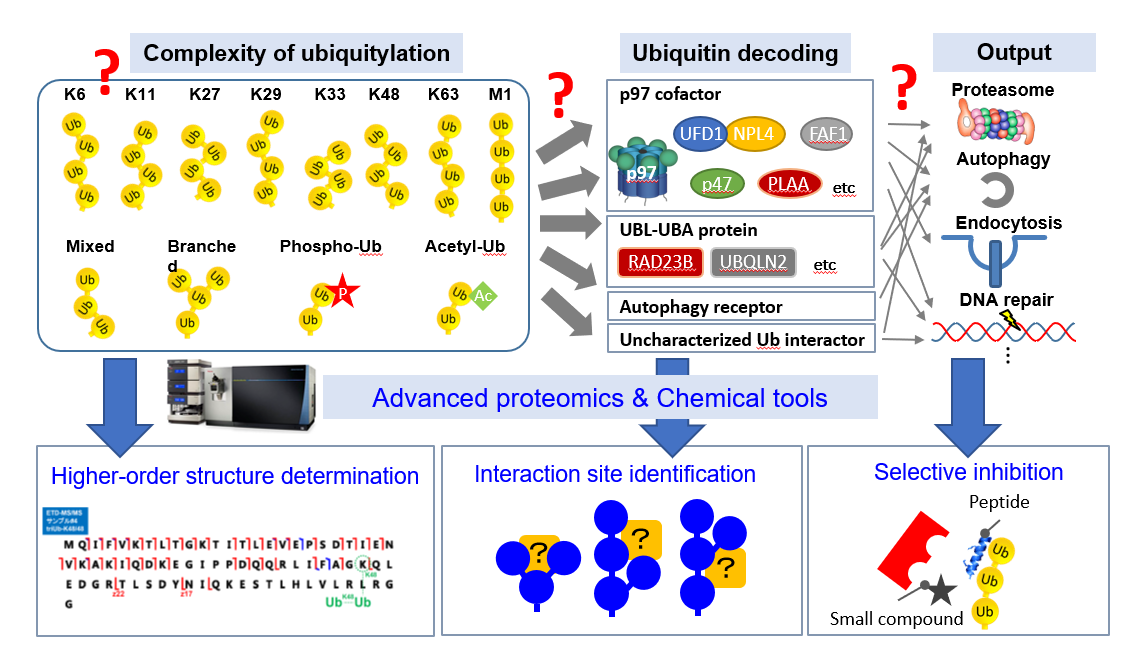
The diverse functions of ubiquitin can be attributed to the structural diversity of ubiquitin modifications with distinct topologies, called the ‘ubiquitin code’. We have contributed to ubiquitin research in Japan by identifying critical ubiquitin chain decoders upstream of proteasomal degradation and developing methods to analyze ubiquitin chain length. However, due to the complexity of ubiquitylation, we do not yet know the overall picture of how the ubiquitin code determines the function and fate of substrate proteins.
In the planned research, we will introduce advanced proteomics methods to this research area, and by a combination with newly developed chemical tools, we will investigate the higher-order architecture of ubiquitin chains, which is the basis of the ubiquitin code, and decoding mechanism by ubiquitin-binding proteins in major ubiquitin-dependent pathways.
1. Development of advanced ubiquitin proteomics methods
Using the most advanced mass spectrometry, we will introduce state-of-the-art proteomics analysis methods to ubiquitin research in Japan. Specifically, we will establish methods such as TMT labeling for comprehensive identification of ubiquitylated substrates, cross-linking analysis for analyzing ubiquitin decoders, and intact mass analysis for higher order structure of ubiquitin chains.
2. Elucidation of the decoding mechanism of the ubiquitin code
We will elucidate the decoding mechanisms for major ubiquitin-binding proteins by a combination of advanced proteomics methods and chemical tools that inhibit the interaction between the ubiquitin chain and the ubiquitin-binding protein. In addition, using chemically synthesized ubiquitin chains, we will search decoder molecules that discriminate higher-order structures of ubiquitin modification.
Publications
- Tomita T, Hirayama S, Sakurai Y, Ohte Y, Yoshihara H, Saeki Y, Hamazaki J, *Murata S.
Specific modification of aged proteasomes revealed by tag-exchangeable knock-in mice.
Mol. Cell. Biol. 39, e00426-18 (2018)
PMID: 30348842 - Ikeuchi K, Tesina P, Matsuo Y, Sugiyama T, Cheng J, Saeki Y, Tanaka K, Becker T, *Beckmann R, *Inada T.
Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways.
EMBO J. 38, e100276 (2019)
PMID: 30609991 - *Masuda Y, Saeki Y, Arai N, Kawai H, Kukimoto I, Tanaka K, *Masutani C.
Stepwise multipolyubiquitantion of p53 by the E6AP-E6 ubiquitin ligase complex.
J. Biol. Chem. 294, 14860-14875 (2019)
PMID: 31492752 - Sato Y, Tsuchiya H, Yamagata A, Okatsu K, Tanaka K, *Saeki Y, *Fukai S.
Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat. Commun. 10, 5708 (2019)
PMID: 31836717
- Yasuda S, Tsuchiya H, Kaiho A, Guo Q, Ikeuchi K, Endo A, Arai N, Ohtake F, Murata S, Inada T, Baumeister W, Fernandez-Busnadiego R, *Tanaka K, *Saeki Y.
Stress- and ubiquitylation-dependent phase separation of the proteasome.
Nature 578, 296-300 (2020)
PMID: 32025036
Highlighted in Cell Res
EurekAlert!
Faculty Opinions - *Nishiyama A, Mulholland C, Bultmann S, Kori A, Endo A, Saeki Y, Qin W, Trummer C, Chiba Y, Yokoyama H, Kumamoto S, Kawakami T, Hojo H, Nagae G, Aburatani H, Tanaka K, *Arita K, *Leonhardt H, *Nakanishi M.
Two distinct modes of DNMT1 recruitment ensure the stable maintenance DNA methylation.
Nat. Commun. 11, 1222 (2020)
PMID: 32144273
Faculty Opinions - Matsuo Y, Tesina P, Nakajima S, Mizuno M, Endo A, Buschauer R, Cheng J, Shounai O, Ikeuchi K, Iwasaki S, Saeki Y, Becker T, *Beckmann R, *Inada T.
RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1.
Nat. Stuct. Mol. Biol. 27, 323-332 (2020)
PMID: 32203490 - Oikawa D, Sato Y, Ohtake F, Komakura K, Hanada K, Sugawara K, Terawaki S, Mizukami Y, Phuong HT, Iio K, Obika S, Fukushi M, Irie T, Tsuruta D, Sakamoto S, Tanaka K, Saeki Y, Fukai S, *Tokunaga F.
Molecular bases for HOIPINs-mediated inhibition of LUBAC and innate immune responses.
Commun. Biol. 3, 163 (2020)
PMID: 32246052 - Fuseya Y, Fujita H, Kim M, Ohtake F, Nishide A, Sasaki K, Saeki Y, Tanaka K, Takahashi R, *Iwai K.
The HOIL-1L ligase modulates immune signalling and cell death via monoubiquitination of LUBAC.
Nat. Cell Biol. 22, 663-673 (2020)
PMID: 32393887 - *Suzuki G, Imura S, Hosokawa M, Katsumata R, Nonaka T, Hisanaga SI, Saeki Y, *Hasegawa M.
α-Synuclein strains that cause distinct pathologies differentially inhibit proteasome.
eLife 9, e56825 (2020)
PMID: 32697196 - *Watanabe M, Saeki Y, Takahashi H, Ohtake F, Yoshida Y, Kasuga Y, Kondo T, Yaguchi H, Suzuki M, Ishida H, Tanaka K, *Hatakeyama S.
A substrate-trapping strategy to find E3 ubiquitin ligase substrates identifies Parkin and TRIM28 targets.
Commun. Biol. 3,592 (2020)
PMID: 33082525 - Matsuki Y, Matsuo Y, Nakano Y, Iwasaki S, Yoko H, Udagawa T, Li S, Saeki Y, Yoshihisa T, Tanaka K, Ingolia N, *Inada T.
Ribosomal protein S7 ubiquitination during ER stress in yeast is associated with selective mRNA translation and stress outcome.
Sci. Rep. 10, 19669 (2020)
PMID: 33184379 - Nakabayashi O, Takahashi H, Moriwaki K, Komazawa-Sakon S, Ohtake F, Murai Y, Koyahara Y, Saeki Y, Yoshida Y, Yamazaki S, Tokunaga F, Sawasaki T, *Nakano H.
MIND bomb 2 prevents RIPK1 kinase activity-dependent and -independent apoptosis through ubiquitylation of cFLIPL.
Commun. Biol. 4, 80 (2021)
PMID: 33469115 - Kaiho-Soma A, Akizuki Y, Igarachi K, Endo A, Shoda T, Kawase Y, Demizu Y, Naito M, Saeki Y, Tanaka K, *Ohtake F.
TRIP12 promotes small molecule-induced degradation of BRD4 through K29/K48 branched ubiquitin chains.
Mol. Cell 81, 1411-1424.e7 (2021)
PMID: 33567268 - Imada T, Shimi T, Kaio A, Saeki Y, *Kimura H.
RNA polymerase II condensate formation and association with Cajal and histone locus bodies in living human cells.
Genes Cells 26, 298-312 (2021)
PMID: 33608942 - Yokoo H, *Shibata N, Endo A, Ito T, Yanase Y, Murakami Y, Fujii K, Hamamura K, Saeki Y, *Naito M, *Aritake K, *Demizu Y.
Discovery of a highly potent and selective PROTAC targeting hematopoietic prostaglandin D synthase via in silico design.
J. Med. Chem. 64, 15868-15882 (2021)
PMID: 34652145 - Huang Y, Yokoe H, Kaiho-Soma A, Takahashi K, Hirasawa Y, Morita H, *Ohtake F, *Kanoh N.
Design, Synthesis, and Evaluation of Trivalent PROTACs Having a Functionalization Site with Controlled Orientation.
Bioconjug Chem. 33, 142-151 (2022)
PMID: 34878263 - Tsunoda T, Riku M, Yamada N, Tsuchiya H, Tomita T, Suzuki M, Kizuki M, Inoko A, Ito H, Murotani K, Murakami H, Saeki Y, *Kasai K.
ENTREP/FAM189A2 downregulated in breast cancer encodes a new activator for ITCH ubiquitin ligase.
EMBO Rep. 23, e51182 (2022)
PMID: 34927784 - Hasegawa Y, Reyes TH, Uemura T, Baral A, Fujimaki A, Luo Y, Morita Y, Saeki Y, Maekawa S, Yasuda S, Mukuta K, Fukao Y, Tanaka K, Nakano A, Takagi J, Bhalerao RP, Yamaguchi J, Sato T.
The TGN/EE SNARE protein SYP61 and the ubiquitin ligase ATL31 cooperatively regulate plant responses to carbon/nitrogen conditions in Arabidopsis.
Plant Cell. 34, 1354-1374 (2022)
PMID: 35089338 - Akizuki Y, Morita M, Mori Y, Kaiho-Soma A, Dixit S, Endo A, Shimogawa M, Hayashi G, Naito M, Okamoto A, Tanaka K, Saeki Y, *Ohtake F.
cIAP1-based degraders induce degradation via branched ubiquitin architectures.
Nat Chem Biol. 19, 311-322 (2022)
PMID: 36316570 - Ishigaki H, Yasui F, Nakayama M, Endo A, Yamamoto N, Yamaji K, Nguyen C, Kitagawa Y, Sanada T, Honda T, Munakata T, Higa M, Toyama S, Kono R, Takagi A, Matsumoto Y, Koseki A, Hayashi K, Shiohara M, Ishii K, Saeki Y, *Itoh Y, *Kohara M.
An attenuated vaccinia vaccine encoding the severe acute respiratory syndrome coronavirus-2 spike protein elicits broad and durable immune responses, and protects cynomolgus macaques and human angiotensin-converting enzyme 2 transgenic mice from severe acute respiratory syndrome coronavirus-2 and its variants.
Front. Microbiol. 13, 967019 (2022)
PMID: 36466631 - Pack CG, Yukii H, Toh-e A, Kudo T, Tsuchiya H, Kaiho A, Sakata E, Murata S, Sako Y, Baumeister W, Tanaka K, *Saeki Y.
Quantitave live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome.
Nat. Commun. 5, 3396 (2014)
PMID: 24598877 - Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe J, Saeki Y, *Tanaka K,*Matsuda N.
Ubiquitin is phosphorylated by PINK1 to activate parkin.
Nature 510, 162-166 (2014)
PMID: 24784582 - *Ohtake F, Saeki Y, Sakamoto K, Ohtake K, Nishikawa H, Tsuchiya H, Ohta T, Tanaka K, Kanno J.
Ubiquitin acetylation inhibits polyubiquitin chain elongation.
EMBO Rep. 16, 192-201 (2015)
PMID: 25527407 - *Yoshida Y, Saeki Y, Murakami A, Kawawaki J, Tsuchiya H, Yoshihara H, Shindo M, *Tanaka K.
A comprehensive method for detecting ubiquitinated substrates using TR-TUBE.
Proc. Natl. Acad. Sci. U.S.A. 112, 4630-4635 (2015)
PMID: 25827227 - *Ohtake F, Saeki Y, Ishido S, Kanno J, *Tanaka K.
The K48-K63 branched ubiquitin chain regulates NF-kB signaling.
Mol. Cell. 64, 251-266 (2016)
PMID: 27746020 - Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K, Ingolia NT, Beckmann R, *Inada T.
Ubiquitination of stalled ribosome triggers ribosome-associated quality control.
Nat. Commun. 8, 159 (2017)
PMID: 28757607 - Sato Y, Okatsu K, Saeki Y, Yamano K, Matsuda N, Kaiho A, Yamagata A, Goto-Ito S, Ishikawa M, Hashimoto Y, Tanaka K, *Fukai S.
Structural basis for specific cleavage of Lys6-linked polyubiquitin chains by USP30.
Nat. Struct. Mol. Biol. 24, 911-919 (2017)
PMID: 28945247 - Tsuchiya H, Ohtake F, Arai N, Kaiho A, Yasuda S, *Tanaka K, *Saeki Y.
In vivo ubiquitin linkage-type analysis reveals that the Cdc48-Rad23/Dsk2 axis contributes to K48-linked chain specificity of the proteasome.
Mol. Cell. 66, 488-502 (2017)
PMID: 28525741 - *Ohtake F, Tsuchiya H, Saeki Y, *Tanaka K.
K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains.
Proc. Natl. Acad. Sci. U.S.A. 115, E1401-E1408 (2018)
PMID: 29378950 - Tsuchiya H, Burana D, Ohtake F, Arai N, Kaiho A, Komada M, *Tanaka K, *Saeki Y.
Ub-ProT reveals global length and composition of protein ubiquitylation in cells.
Nat. Commun. 9, 524 (2018)
PMID: 29410401