Research Group
TeamResearch Group
Research group A02: Development of ubiquitin chemo-technologies
Chemical synthesis of ubiquitin codes
Akimitsu Okamoto, PhDDepartment of Chemistry and Biotechnology, Graduate School of Engineering, The University of Tokyo |
We will chemically synthesize ubiquitin codes and super-ubiquitins and use them to elucidate the mechanism of ubiquitin code function. First, we will establish a method for the chemical synthesis of ubiquitin and apply it to the synthesis of various ubiquitin chains and artificially functional ubiquitins. Next, we will use the synthetic ubiquitin chain to analyze the function of the decoder molecule and analyze the structure and dynamics of the ubiquitin chains.
1. Establishment of chemical synthesis method of ubiquitin
Ubiquitins are divided into two or three fragments, and they are chemically synthesized by preparing those fragment peptides, linking the fragments by chemical ligation, and desulfurizing the cysteines used for ligation.
2. Combinatorial chemical synthesis of various ubiquitin chains
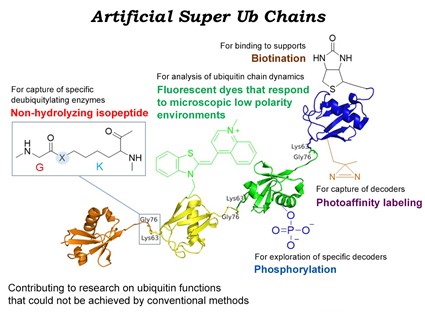
By preparing in advance a fragment into which special ε-Lys-Gly has been appropriately introduced, various polyubiquitin chains can be chemically synthesized in a combinatorial manner. Next, we will start synthesizing phosphorylated ubiquitin chains, acetylated ubiquitin chains, and complex ubiquitin-encoded K48/K63 branched chains and K63/M1 mixed chains. Specific decoder molecules will be captured using a chemically synthesized ubiquitin chain as a bait, and identified using a state-of-the-art mass spectrometer.
3. Synthesis of artificially functional ubiquitin (super-ubiquitin)
We produce ubiquitins into which a fluorescent dye or photoaffinity label has been introduced site-specifically. In addition, we will examine the syntheses of ubiquitins having a biotin-modified end and ubiquitins resistant to deubiquitinase. The created super-ubiquitin will be introduced into cells, and intracellular localization and cellular response will be analyzed.
4. Analysis of properties of super-ubiquitin chains and formations of ubiquitin codes
We will examine the structural analysis and binding analysis of synthetic polyubiquitin chains, and develop a system to monitor ubiquitin code formation using the ubiquitin emitting fluorescence in a specific environment.
5. Ubiquitin single molecule observation
By introducing two different fluorescent labels into polyubiquitin and using fluorescence cross-correlation spectroscopy, we can examine the behavior of ubiquitin at the molecular level in vitro and in cells and their binding to other molecules.
Publications
- Sueoka T, Koyama K, Hayashi G, *Okamoto A.
Chemistry-driven Epigenetic Investigation of Histone and DNA Modifications.
Chem. Rec. 18, 1727-1744 (2018)
PMID: 30070422 - Kamo N, *Hayashi G, *Okamoto A.
Triple Function of 4-Mercaptophenylacetic Acid Promotes One-Pot Multiple Peptide Ligation.
Angew. Chem. Int. Ed. 57, 16533-16537 (2018)
PMID: 30346110 - *Hayashi G, Yanase M, Nakatsuka Y, *Okamoto A.
Simultaneous and Traceless Ligation of Peptide Fragments on DNA Scaffold.
Biomacromolecules 20, 1246-1253 (2019)
PMID: 30677290 - Yanase M, Nakatsu K, Cardos C J, Konda Y, *Hayashi G, *Okamoto A.
Cysteinylprolyl Imide (CPI) Peptide: A Highly Reactive and Easily Accessible Crypto-thioester for Chemical Protein Synthesis.
Chem. Sci. 10, 5967-5975 (2019)
PMID: 31360403 - Kamo N, *Hayashi G, *Okamoto A.
Chemical Synthesis of Cys-Containing Protein via Chemoselective Deprotection with Different Palladium Complexes.
Org. Lett. 21, 8378-8382 (2019)
PMID: 31560553 - Izuta S, *Yamaguchi S, Kosaka T, *Okamoto A.
Reversible and Photoresponsive Immobilization of Nonadherent Cells by Spiropyran-Conjugated PEG-Lipids.
ACS Appl. Bio Mater. 2, 33-38 (2019) - *Yamaguchi S, Higashi K, Azuma T, *Okamoto A.
Supramolecular Polymeric Hydrogels for Ultrasound-guided Protein Release.
Biotechnol. J. 14, 1800530 (2019)
PMID: 30810275 - Usami K, Xiao K, *Okamoto A.
Efficient Ketose Production by Hydroxyapatite Catalyst in a Continuous Flow Module.
ACS Sustainable Chem. Eng. 7, 3372-3377 (2019) - Jarzebska N A, *Yamaguchi S, Izuta S, Kosaka T, Yamahira S, Nagamune T, *Okamoto A.
Photo-responsive materials with strong cell trapping ability for light-guided manipulation of nonadherent cells.
Biomater. Sci. 7, 4514-4518 (2019)
PMID: 31602442 - Morihiro K, *Okamoto A.
A highly constrained nucleic acid analog based on α-L-threosamine.
Nucleosides Nucleotides Nucleic Acids 39, 270-279 (2020)
PMID: 31530088 - Chen J, Morihiro K, Fukui D, Guo L, *Okamoto A.
Live‐Cell Sensing of Telomerase Activity Using Hybridization-Sensitive Fluorescent Oligonucleotide Probes.
ChemBioChem 21, 1022-1027 (2020)
PMID: 31840916 - Nakatsu K, Yanase M, *Hayashi G, *Okamoto A.
Fmoc-Compatible and C-terminal-Sequence-Independent Peptide Alkyl Thioester Formation Using Cysteinylprolyl Imide.
Org. Lett. 22, 4670–4674 (2020)
PMID: 32484687 - *Yamaguchi S, Takagi R, Hosogane T, Ohashi Y, Sakai Y, Sakakihara S, Iino R, Tabata K, Noji H, *Okamoto A.
Single cell array enclosed with a photodegradable hydrogel in microwells for image-based cell classification and selective photorelease of cells.
ACS Appl. Bio Mater. 3, 5887–5895 (2020) - Ogata M, Hayashi G, Ichiu A, *Okamoto A.
L-DNA-tagged fluorescence in situ hybridization for highly sensitive imaging of RNAs in single cells.
Org. Biomol. Chem. 18, 8084-8088 (2020)
PMID: 33001106 - Hao F, Murphy KJ, Kujirai T, Kamo N, Kato J, Koyama M, Okamoto A, Hayashi G, Kurumizaka H, *Hayes JJ.
Acetylation-Modulated Communication between the H3 N-Terminal Tail Domain and the Intrinsically Disordered H1 C-Terminal Domain.
Nucleic Acids Res. 48, 11510–11520 (2020)
PMID: 33125082 - Morihiro K, Ishinabe T, Takatsu M, Osumi H, Osawa T, *Okamoto A.
Floxuridine Oligomers Activated under Hypoxic Environment.
J. Am. Chem. Soc. 143, 3340–3347 (2021)
PMID: 33648338 - Kamo N, Kujirai T, Kurumizaka H, Murakami H, *Hayashi G, *Okamoto A.
Organoruthenium-Catalyzed Chemical Protein Synthesis to Elucidate the Functions of Epigenetic Modifications on Heterochromatin Factors.
Chem. Sci. 12, 5926-5937 (2021) - Kamo N, *Hayashi G, *Okamoto A.
Silyl-Protected Propargyl Glycine for Multiple Labeling of Peptides by Chemoselective Silyl-Deprotection.
Tetrahedron Lett. 73, 153093 (2021) - Koyama K, Hayashi G, Ueda H, Ota S, Nagae G, Aburatani H, *Okamoto A.
Base-Resolution Analysis of 5-Hydroxymethylcytidine by Selective Oxidation and Reverse Transcription Arrest.
Org. Biomol. Chem. 19, 6478-6486 (2021)
PMID: 34241617 - Morihiro K, Moriyama Y, Nemoto Y, Osumi H, *Okamoto A.
anti-syn Unnatural Base Pair Enables Alphabet-Expanded DNA Self-Assembly.
J. Am. Chem. Soc. 143, 14207-14217 (2021) - Pan M, Zorbas C, Sugaya M, Ishiguro K, Kato M, Nishida M, Zhang H-F, Candeias M M, Okamoto A, Ishikawa T, Soga T, Aburatani H, Sakai J, Matsumura Y, Suzuki T, Proud C G, Lafontaine D L J, *Osawa T.
Glutamine Deficiency in Solid Tumor Cells Confers Resistance to Ribosomal RNA Synthesis Inhibitors.
Nat. Commun. 13, 3706 (2022)
PMID: 35764642 - Yamahira S, Misawa R, Kosaka T, Tan M, Izuta S, Yamashita H, Heike Y, Okamoto A, Nagamune T, *Yamaguchi S.
Photoactivatable Materials for Versatile Single-Cell Patterning Based on the Photocaging of Cell-Anchoring Moieties through Lipid Self-Assembly.
J. Am. Chem. Soc. 144, 13154-13162 (2022)
PMID: 35767880 - Nakatsu K, Okamoto A, Hayashi G, *Murakami H.
Repetitive Thiazolidine Deprotection Using a Thioester-Compatible Aldehyde Scavenger for One-Pot Multiple Peptide Ligation.
Angew. Chem. Int. Ed. 61, e202206240 (2022)
PMID: 35881031 - *Yamaguchi S, Ikeda R, Umeda Y, Kosaka T, Yamahira S, *Okamoto A.
Chemoenzymatic Labeling to Visualize Intercellular Contacts Using Lipidated Sortase A.
ChemBioChem 23, e202200474 (2022)
PMID: 35976800 - Kosaka T, *Yamaguchi S, Izuta S, Yamahira S, Shibasaki Y, Tateno H, *Okamoto A.
Bioorthogonal Photoreactive Surfaces for Single-Cell Analysis of Intercellular Communications.
J. Am. Chem. Soc. 144, 17980-17988 (2022)
PMID: 36126284 - *Yamaguchi S, Yamamoto K, Yamamoto R, Takamori S, Ishiwatari A, Minamihata K, Nagamune T, *Okamoto A.
Intracellular Protein Photoactivation Using Sterically Bulky Caging.
ChemBioChem 23, e202200476 (2022)
PMID: 36173993 - Akizuki Y, Morita M, Mori Y, Kaiho-Soma A, Dixit S, Endo A, Shimogawa M, Hayashi G, Naito M, Okamoto A, Tanaka K, Saeki Y, *Ohtake F.
cIAP1-based degraders induce degradation via branched ubiquitin architectures.
Nat Chem Biol. 19, 311-322 (2023)
PMID: 36316570 - *Morihiro K, Osumi H, Morita S, Hattori T, Baba M, Harada N, Ohashi R, *Okamoto A.
Oncolytic Hairpin DNA Pair: Selective Cytotoxic Inducer through MicroRNA-Triggered DNA Self-Assembly.
J. Am. Chem. Soc. 145, 135-142 (2023)
PMID: 36538570 - Liu S, Morihiro K, Takeuchi F, Li Y, *Okamoto A.
Interstrand Crosslinking Oligonucleotides Elucidate the Effect of Metal Ions on the Methylation Status of Repetitive DNA Elements.
Front. Chem. 11, 1122474 (2023)
PMID: 36711237
Former Publications
- *Okamoto A.
DNA-Osmium Complexes: Recent Developments in the Operative Chemical Analysis of DNA Epigenetic Modification.
ChemMedChem 9, 1958-1965 (2014)
PMID: 24989699 - Hayashi G, Sakamoto R, *Okamoto A.
2-Oxazoline Formation for Selective Chemical Labeling of 5-Hydroxylysine.
Chem. Asian J. 10, 1138-1141 (2015)
PMID: 25757225 - Yamamoto K, Chikaoka Y, Hayashi G, Sakamoto R, Yamamoto R, Sugiyama A, Kodama T, Okamoto A, *Kawamura T.
Middle-Down and Chemical Proteomic Approaches to Reveal Histone H4 Modification Dynamics in Cell Cycle: Label-Free Semi-Quantification of Histone Tail Peptide Modifications Including Phosphorylation and Highly Sensitive Capture of Histone PTM Binding Proteins Using Photo-Reactive Crosslinkers.
Mass Spectrom. 4, A0039 (2015)
PMID: 26819910 - Hayashi G, Sueoka T, *Okamoto A.
In vitro and in cell analysis of chemically synthesized histone H2A with multiple modifications.
Chem. Commun. 52, 4999-5002 (2016)
PMID: 26981611 - Hayashi G, Koyama K, Shiota H, Kamio A, Umeda T, Nagae G, Aburatani H, *Okamoto A.
Base-Resolution Analysis of 5-Hydroxymethylcytosine by One-Pot Bisulfite-Free Chemical Conversion with Peroxotungstate.
J. Am. Chem. Soc. 138, 14178-14181 (2016)
PMID: 27766835 - Yamaguchi S, Matsushita T, Izuta S, Katada S, Ura M, Ikeda T, Hayashi G, Suzuki Y, Kobayashi Y, Tokunaga K, Ozeki Y, *Okamoto A.
Chemically-activatable alkyne-tagged probe for imaging microdomains in lipid bilayer membranes.
Sci. Rep. 7, 41007 (2017)
PMID: 28117375 - Matsushita T, Moriyama Y, Nagae G, Aburatani H, *Okamoto A.
DNA-friendly Cu(II)/TEMPO-catalyzed 5-hydroxymethylcytosine-specific oxidation.
Chem. Commun. 53, 5756-5759 (2017)
PMID: 28492666 - Hayashi G, Kamo N, *Okamoto A.
Chemical synthesis of dual labeled protein via differently protected alkynes enables intramolecular FRET analysis.
Chem. Commun. 53, 5918-5921 (2017)
PMID: 28513662 - Guo L, *Okamoto A.
Fluorescence-switching RNA for detection of bacterial ribosome.
Chem. Commun. 53, 9406-9409 (2017)
PMID: 28765840 - Sueoka T, Hayashi G, *Okamoto A.
Regulation of the Stability of the Histone H2A-H2B Dimer by H2A Tyr57 Phosphorylation.
Biochemistry 56, 4767-4772 (2017)
PMID: 28813589