Research Group
TeamResearch Group
Research group A01: Elucidation of ubiquitin functions by chemo-technologies
Regulation of protein degradation using chemo-technologies
Shigeo Murata. MD, PhDThe University of Tokyo, Graduate School of Pharmaceutical Sciences |
|
Koji Yamano, PhDDepartment of Biomolecular Pathogenesis, Medical Research Institute,
Tokyo Medical and Dental University |
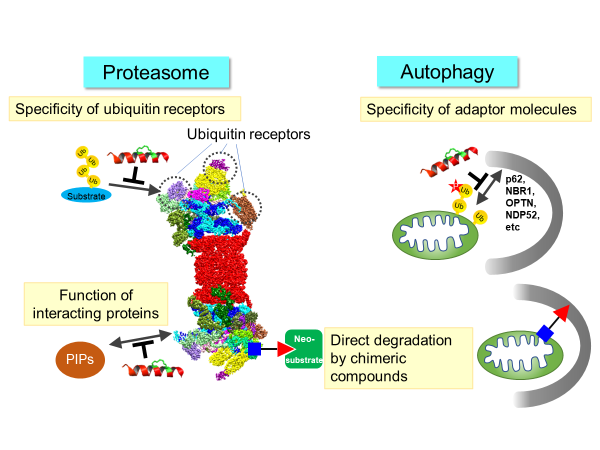
Proteasomes and autophagy are the main degradation systems in eukaryotic cells and play a central role in the dynamic metabolism of proteins and organelles that support vital activities. In both degradation systems, ubiquitin modification and its receptor molecules play an essential role in selecting a target to be degraded. This study focuses on proteolysis and organelle degradation, one of the primary roles of ubiquitin, and uses compounds and membrane-permeable peptides capable of timely losing specific protein-protein interactions. We aim to elucidate a new understanding of substrate selectivity and regulatory mechanism for intracellular degradation that are hard to be solved by gene manipulation. Furthermore, we aim to create chimeric compounds that directly link both degradation systems and degradation targets and aim to establish a methodology for directly and selectively inducing protein degradation without involving ubiquitination.
Publications
- Okatsu K, Sato Y, Yamano K, Matsuda N, Negishi L, Takahashi A, Yamagata A, Goto-Ito S, Mishima M, Ito Y, Oka T, Tanaka K, *Fukai S.
Structural insights into ubiquitin phosphorylation by PINK1.
Sci. Rep. 8, 10382 (2018)
PMID: 29991771 - Wu W, Sahara K, Hirayama S, Zhao X, Watanabe A, Hamazaki J, Yashiroda H, *Murata S.
PAC1‐PAC2 proteasome assembly chaperone retains the core α4–α7 assembly intermediates in the cytoplasm.
Genes to Cells 23, 839–848 (2018)
PMID: 30133132 - Tomaru U, Konno S, Miyajima S, Kimoto R, Onodera M, Kiuchi S, Murata S, Ishizu A, Kasahara M.
Restricted Expression of the Thymoproteasome Is Required for Thymic Selection and Peripheral Homeostasis of CD8+ T Cells.
Cell Rep. 26, 639-651.e2 (2019)
PMID: 30650357 - Ohigashi I, Tanaka Y, Kondo K, Fujimori S, Kondo H, Palin AC, Hoffmann V, Kozai M, Matsushita Y, Uda S, Motosugi R, Hamazaki J, Kubota H, Murata S, Tanaka K, Katagiri T, Kosako H, Takahama Y.
Trans-omics Impact of Thymoproteasome in Cortical Thymic Epithelial Cells.
Cell Rep. 29, 2901-2916.e6 (2019)
PMID: 31775054 - Otsubo R, Mimuro H, Ashida H, Hamazaki J, Murata S, Sasakawa C.
Shigella effector IpaH4.5 targets 19S regulatory particle subunit RPN13 in the 26S proteasome to dampen cytotoxic T lymphocyte activation.
Cell Microbiol. 21, e12974 (2019)
PMID: 30414351 - Bai M, Zhao X, Sahara K, Ohte Y, Hirano Y, Kaneko T, Yashiroda H, Murata S.
In-depth Analysis of the Lid Subunits Assembly Mechanism in Mammals.
Biomolecules 9, 213 (2019)
PMID: 31159305 - Arata Y, Watanabe A, Motosugi R, Iemura S, Natsume T, Mukai K, Taguchi T, Hirayama S, Hamazaki J, Murata S.
FAM48A mediates compensatory autophagy induced by proteasome impairment.
Genes to Cells 24, 559–568 (2019)
PMID: 31210371 - Koyano F, Yamano K, Kosako H, Tanaka K, *Matsuda N.
Parkin recruitment to impaired mitochondria for nonselective ubiquitylation is facilitated by MITOL.
J. Biol. Chem. 294, 10300-10314 (2019)
PMID: 31110043 - Koyano F, Yamano K, Kosako H, Kimura Y, Kimura M, Fujiki Y, Tanaka K, *Matsuda N.
Parkin-mediated ubiquitylation redistributes MITOL/March5 from mitochondria to peroxisomes.
EMBO Rep. 20, e47728 (2019)
PMID: 31602805 - Arata Y, Watanabe A, Motosugi R, Murakami R, Goto T, Hori S, Hirayama S, Hamazaki J, Murata S.
Defective induction of the proteasome associated with T‐cell receptor signaling underlies T‐cell senescence.
Genes to Cells 24, 801–813 (2019)
PMID: 31621149 - Yasuda S, Tsuchiya H, Kaiho A, Guo Q, Ikeuchi K, Endo A, Arai N, Ohtake F, Murata S, Inada T, Baumeister W, Fernández-Busnadiego R, Tanaka K, *Saeki Y.
Stress- and ubiquitylation-dependent phase separation of the proteasome.
Nature 578, 296–300 (2020)
PMID: 32025036
Highlighted in Cell Res
EurekAlert!
Faculty Opinions - Waku T, Nakamura N, Koji M, Watanabe H, Katoh H, Tatsumi C, Tamura N, Hatanaka A, Hirose S, Katayama H, Tani M, Kubo Y, Hamazaki J, Hamakubo T, Watanabe A, Murata S, Kobayashi A.
NRF3-POMP-20S Proteasome Assembly Axis Promotes Cancer Development via Ubiquitin-Independent Proteolysis of p53 and Retinoblastoma Protein.
Mol. Cell. Biol. 40, e597-19 (2020)
PMID: 32123008 - *Yamano K, Kikuchi R, Kojima W, Hayashida R, Koyano F, Kawawaki J, Shoda T, Demizu Y, Naito M, Tanaka K, *Matsuda N.
Critical role of mitochondrial ubiquitination and the OPTN–ATG9A axis in mitophagy.
J. Cell Biol. 219, e201912144 (2020)
PMID: 32556086 - Hashimoto E, Okuno S, Hirayama S, Arata Y, Goto T, Kosako H, Hamazaki J, *Murata S.
Enhanced O-GlcNAcylation mediates cytoprotection under proteasome impairment by promoting proteasome turnover in cancer cells.
iScience 23, 101299 (2020)
PMID: 32634741 - Kojima W, *Yamano K, Kosako H, Imai K, Kikuchi R, Tanaka K, Matsuda N.
Mammalian BCAS3 and C16orf70 associate with the phagophore assembly site in response to selective and non-selective autophagy.
Autophagy 17, 2011-2036 (2021)
PMID: 33499712 - Takehara Y, Yashiroda H, Matsuo Y, Zhao X, Kamigaki A, Matsuzaki T, Kosako H, Inada T, *Murata S.
The ubiquitination-deubiquitination cycle on the ribosomal protein eS7A is crucial for efficient translation.
iScience 24, 102145 (2021)
PMID: 33665564 - *Kanazawa N, Hemmi H, Kinjo N, Ohnishi H, Hamazaki J, Mishima H, Kinoshita A, Mizushima T, Hamada S, Hamada K, Kawamoto N, Kadowaki S, Honda Y, Izawa K, Nishikomori R, Tsumura M, Yamashita Y, Tamura S, Orimo T, Ozasa T, Kato T, Sasaki I, Fukuda-Ohta Y, Wakaki-Nishiyama N, Inaba Y, Kunimoto K, Okada S, Taketani T, Nakanishi K, Murata S, Yoshiura K, *Kaisho T.
Heterozygous missense variant of the proteasome subunit β-type 9 causes neonatal-onset autoinflammation and immunodeficiency.
Nature Comm.12, 6819 (2021)
PMID: 34819510 - Watanabe A, Hama K, Watanabe K, Fujiwara Y, Yokoyama K, Murata S, *Takita R.
Controlled tetradeuteration of straight-chain fatty acids: synthesis, application, and insight into the metabolism of oxidized linoleic acid.
Angew Chem Int Ed Engl. e202202779 (2022)
PMID: 35411582 - *Tanahashi N, Komiyama M, Tanaka M, Yokobori Y, Murata S, Tanaka K.
The effect of nutrient deprivation on proteasome activity in 4-week-old mice and 24-week-old mice.
J Nutr Biochem 105, 108993 (2022)
PMID: 35331898 - Hayashida R, Kikuchi R, Imai K, Kojima W, Yamada T, Iijima M, Sesaki H, Tanaka K, *Matsuda N, *Yamano K.
Elucidation of ubiquitin-conjugating enzymes that interact with RBR-type ubiquitin ligases using a liquid-liquid phase separation-based method.
J Biol Chem. 299, 102822 (2023)
PMID: 36563856
Former Publications
- Yamano K, Fogel AI, Wang C, van der Bliek, AM, Youle RJ.
Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy.
eLife 3, e01612 (2014)
PMID: 24569479 - Hamazaki J, Hirayama S, *Murata S.
Redundant roles of Rpn10 and Rpn13 in recognition of ubiquitinated proteins and cellular homeostasis.
PLoS Genet. 11, e1005401 (2015)
PMID: 26222436 - Yamano K, Queliconi BB, Koyano F, Saeki Y, Hirokawa T, Tanaka K, Matsuda N.
Site-specific interaction mapping of phosphorylated ubiquitin to uncover Parkin activation.
J. Biol. Chem. 290, 25199-25211 (2015)
PMID: 26260794 - Sasaki K, Takada K, Ohte Y, Kondo H, Sorimachi H, Tanaka K, *Takahama Y, *Murata S.
Thymoproteasomes produce unique peptide motifs for positive selection of CD8+ T cells.
Nat. Commun. 6, 7484 (2015)
PMID: 26099460 - Yamano K, Matsuda N, Tanaka K.
The Ubiquitin signal and autophagy: an orchestrated dance leading to mitochondrial degradation.
EMBO Rep. 17, 300-316 (2016)
PMID: 26882551 - Koizumi S, Irie T, Hirayama S, Sakurai Y, Yashiroda H, Naguro I, Ichijo H, Hamazaki J, *Murata S.
The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction.
elife 5, e18357 (2016)
PMID: 27528193 - Lu X, Nowicka U, Sridharan V, Liu F, Randles L, Hymel D, Dyba M, Tarasov SG, Tarasova NI, Zhao XZ, Hamazaki J, Murata S, Burke TR Jr, *Walters KJ.
Structure of the Rpn13-Rpn2 complex provides insights for Rpn13 and Uch37 as anticancer targets.
Nat. Commun. 8, 15540 (2017)
PMID: 28598414 - Yamano K, Wang C, Sarraf SA, Münch C, Kikuchi R, Noda NN, Hizukuri Y, Kanemaki MT, Harper W, Tanaka K, Matsuda N, Youle RJ.
Endosomal Rab cycles regulate Parkin-mediated mitophagy.
eLife 7, e31326 (2018)
PMID: 29360040 - Uechi H, Kuranaga E, Iriki T, Takano K, Hirayama S, Miura M, Hamazaki J, *Murata S.
Ubiquitin-Binding Protein CG5445 Suppresses Aggregation and Cytotoxicity of Amyotrophic Lateral Sclerosis-linked TDP-43 in Drosophila.
Mol. Cell. Biol. 38, e00195-17 (2018)
PMID: 29109084 - *Hirayama S, Sugihara M, Morito D, Iemura S, Natsume T, Murata S, *Nagata K.
Nuclear export of ubiquitinated proteins via the UBIN-POST system.
Proc. Natl. Acad. Sci. U.S.A. 115, E4199-E4208 (2018)
PMID: 29666234 - Murata S, Takahama Y, Kasahara M, Tanaka K.
The immunoproteasome and thymoproteasome: functions, evolution and human disease.
Nat. Immunol. 19,923-931 (2018)
PMID: 30104634